For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAVKVASSWINGASVETSGETHQIIDPATGKSVSEYNLATAADVDTAVAAARAAFPAWAGATPAERSAVL AKLAKLADEHAEQFVAEEVSQTGKPVRLATEFDVPGSVDNIDFFAGAARHLEGKATAEYSGDHTSSIRRE AVGVVATITPWNYPLQMAVWKVIPALAAGCAVVIKPAEITPLTTLTLARLASEAGLPDGVFNVVTGSGAE VGTALAGHRDVDVVTFTGSTAVGRKVMASAAVHGHRTQLELGGKAPFVVFSDADLDAAVQGAVAGALINT GQDCTAATRAIVARELYDDFVAGVAEVMGKIVVGDPHDPDTDLGPLISYAHRDKVAGMVARAPQQGGRVV TGGEAPDLPGAFYRPTLIADVDESSEVYRDEIFGPVLTVRSFTDDDDALRQANDTEYGLAASAWTRDVYR AQRASREINAGCVWINDHIPIISEMPHGGVGASGFGKDMSDYSFEEYLTIKHVMSDITGVAEKEWHRTIF NKR
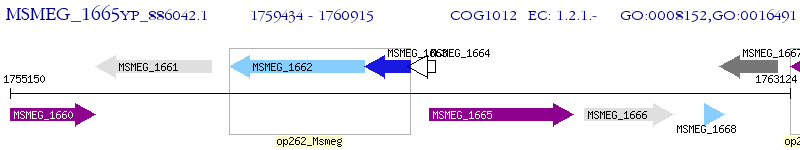
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1665 | - | - | 100% (493) | gamma-aminobutyraldehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6297 | - | e-103 | 43.40% (477) | aldehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2597 | - | 1e-91 | 42.79% (437) | aldehyde dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2883c | aldC | 3e-89 | 40.62% (453) | aldehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4860 | - | 0.0 | 85.28% (489) | gamma-aminobutyraldehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv2858c | aldC | 3e-89 | 40.62% (453) | aldehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 7e-71 | 36.59% (451) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3328 | - | 0.0 | 81.26% (491) | gamma-aminobutyraldehyde dehydrogenase |
| M. marinum M | MMAR_3556 | - | 0.0 | 79.43% (491) | aldehyde dehydrogenase NAD dependent |
| M. avium 104 | MAV_2608 | - | 1e-98 | 42.43% (469) | aldehyde dehydrogenase family protein |
| M. thermoresistible (build 8) | TH_2957 | - | 9e-85 | 39.61% (462) | PUTATIVE aldehyde dehydrogenase |
| M. ulcerans Agy99 | MUL_2789 | - | 0.0 | 78.00% (491) | gamma-aminobutyraldehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1572 | - | 0.0 | 85.69% (489) | gamma-aminobutyraldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2883c|M.bovis_AF2122/97 -------------------------MSTTQLINPATEEVLASVDHTDANA
Rv2858c|M.tuberculosis_H37Rv -------------------------MSTTQLINPATEEVLASVDHTDANA
Mflv_4860|M.gilvum_PYR-GCK --------MTVASSWIDGAPVKTG-GAQHTVINPATGESVAEFALAQPGD
Mvan_1572|M.vanbaalenii_PYR-1 ---------MLAGSWIDGAPVNTR-GPVHQVINPATGAPVAEYALAQPAD
MSMEG_1665|M.smegmatis_MC2_155 ------MAVKVASSWINGASVETS-GETHQIIDPATGKSVSEYNLATAAD
MAB_3328|M.abscessus_ATCC_1997 --------MSVPSSWINGRPVATH-GDTHRVINPATGEAVADLKLATTAD
MMAR_3556|M.marinum_M ----MTTTVSVAGSWINGAAHSTG-GRQHAVINPATQETIAELALATPAD
MUL_2789|M.ulcerans_Agy99 ----MTTTVSVAGSWINGAAHSTG-GRQHAVINPATQETIAELALATPAD
MAV_2608|M.avium_104 MTDTATETLVQFDLLIGGESSSAASGLTYDSVDPFTGRPWARVPNAAAAD
TH_2957|M.thermoresistible__bu MTSTLER--PEVTLRIGGEKRRTASGGTFDHINPCTGAPDAAIPLAGPAE
MLBr_02573|M.leprae_Br4923 --------------------------MSIATINPATGETVKTFTPTTQDE
::* * :
Mb2883c|M.bovis_AF2122/97 VDDAVQRARAAQRR-WARLAPAQRAAGLRAFAAAVQAHLDELAALEVANS
Rv2858c|M.tuberculosis_H37Rv VDDAVQRARAAQRR-WARLAPAQRAAGLRAFAAAVQAHLDELAALEVANS
Mflv_4860|M.gilvum_PYR-GCK VDRAVVSARAALPG-WKGATPAERSAVLAKLAKLADDATADLVAEEVSQT
Mvan_1572|M.vanbaalenii_PYR-1 VDAAVASARAALGG-WATATPAERSAVLAKLAKLADEATADLVAEEVSQT
MSMEG_1665|M.smegmatis_MC2_155 VDTAVAAARAAFPA-WAGATPAERSAVLAKLAKLADEHAEQFVAEEVSQT
MAB_3328|M.abscessus_ATCC_1997 VDAAVAAARAAGPA-WASSTPVDRSSVLAKLAAVLSANADTLVADEVAQT
MMAR_3556|M.marinum_M VDRAVVAARRALPQ-WAGVTPAERSAVLAKLADLLEQRAPELIAEEVCQT
MUL_2789|M.ulcerans_Agy99 VDHAVVAARRALPQ-WAGVTTAERSAVLAELADLLEQRAPELIAEEVCQT
MAV_2608|M.avium_104 VDRAVGAARAALNGPWGALTATARGKLLHRLGEIIAREADQLAELEVRDG
TH_2957|M.thermoresistible__bu IDDAVKAAHDAFHS-WRQTNPAERRRLLFRLAELIEQHTDDFVRLGTFDN
MLBr_02573|M.leprae_Br4923 VEDAIARAYARFDN-YRHTTFTQRAKWANATADLLEAEANQSAALMTLEM
:: *: * : . * . . :
Mb2883c|M.bovis_AF2122/97 GHPI-VSAEWEAGHVRDVLAFYAASPERLSGRQIPVA---GGVDVTFNEP
Rv2858c|M.tuberculosis_H37Rv GHPI-VSAEWEAGHVRDVLAFYAASPERLSGRQIPVA---GGVDVTFNEP
Mflv_4860|M.gilvum_PYR-GCK GKPVRLATEFDVPGSVDNIDFFAGAARHLEGKASAEY-SGDHTSSIRREA
Mvan_1572|M.vanbaalenii_PYR-1 GKPVRLATEFDVPGSVDNIDFFAGAARHLEGKASGEY-SADHTSSIRREA
MSMEG_1665|M.smegmatis_MC2_155 GKPVRLATEFDVPGSVDNIDFFAGAARHLEGKATAEY-SGDHTSSIRREA
MAB_3328|M.abscessus_ATCC_1997 GKPVRLASEFDVPGSIDNVEFFAGAARHLEGKATAEY-SADHTSSIRREA
MMAR_3556|M.marinum_M GKPLRLAEEFDVPGSVDNIRFFAGAARRLPGQATAEY-SADHTSSIRREA
MUL_2789|M.ulcerans_Agy99 GKPLRLAEEFDVPGSVDNIRFFAGAARRLPGQATAEY-SADHTSSIRREA
MAV_2608|M.avium_104 GKLVREMVG-QMRSLPDYYFYYGGLADKLQGDVIPVD-KPNYFVYTRHEP
TH_2957|M.thermoresistible__bu GTPVGLVAG-LVAMSAEWTRYYAGWADKITSEVSGTYGTTGEFSYTLSQP
MLBr_02573|M.leprae_Br4923 GKTLQSAKDEAMKCAKGFRYYAEKAETLLADEPAEAAAVGASKALARYQP
* : : : . :.
Mb2883c|M.bovis_AF2122/97 MGVVGVITPWNFPMVIASWAIAPALAAGNAVLVKPAELTPLTTMRLGELA
Rv2858c|M.tuberculosis_H37Rv MGVVGVITPWNFPMVIASWAIAPALAAGNAVLVKPAELTPLTTMRLGELA
Mflv_4860|M.gilvum_PYR-GCK VGVVATITPWNYPLQMAVWKVIPALAAGCSVVIKPAEITPLTTLTLARLA
Mvan_1572|M.vanbaalenii_PYR-1 VGVVATITPWNYPLQMAVWKVLPALAAGCSVVIKPAEITPLTTLTLARLA
MSMEG_1665|M.smegmatis_MC2_155 VGVVATITPWNYPLQMAVWKVIPALAAGCAVVIKPAEITPLTTLTLARLA
MAB_3328|M.abscessus_ATCC_1997 AGVVATITPWNYPLQMAVWKVLPALAAGCTVVIKPSELTPLTTLTLARLA
MMAR_3556|M.marinum_M IGVVATITPWNYPLQMAVWKVMPALAAGCTVVIKPSELTPLTTLTLARLA
MUL_2789|M.ulcerans_Agy99 IGVVATITPWNYPLQMAVWKVMPALAAGCTVVIKPSELTPLTTLTLARLA
MAV_2608|M.avium_104 VGVVGAITPWNSPLLLLTWKLAAGLAAGCTFVVKPSDHTPTSTLAFAKLF
TH_2957|M.thermoresistible__bu YGVIGVIITWNGPLISLAMKVPAALAAGNTVVVKPSELTPFSATLFADLA
MLBr_02573|M.leprae_Br4923 LGVVLAVMPWNFPLWQTVRFAAPALMAGNVGLLKHASNVPQCALYLADVI
**: .: .** *: ..* ** ::* :. .* : :. :
Mb2883c|M.bovis_AF2122/97 VEAGLDEDLLQVLPGKGTVVGERFVTHPDIRKIVFTGSTEVGKRVMAGAA
Rv2858c|M.tuberculosis_H37Rv VEAGLDEDLLQVLPGKGTVVGERFVTHPDIRKIVFTGSTEVGKRVMAGAA
Mflv_4860|M.gilvum_PYR-GCK SEAGLPDGVLNVVTGSGADVGAALAGHADVDMVTFTGSTPVGRRVMLAAA
Mvan_1572|M.vanbaalenii_PYR-1 AEAGLPAGVFNVVTGSGADVGTALAGHADVDMVTFTGSTPVGRKVMLAAA
MSMEG_1665|M.smegmatis_MC2_155 SEAGLPDGVFNVVTGSGAEVGTALAGHRDVDVVTFTGSTAVGRKVMASAA
MAB_3328|M.abscessus_ATCC_1997 AEAGLPEGVLNVVTGRGDDVGYALASHPDVDLVTFTGSTAVGRKVMAAAA
MMAR_3556|M.marinum_M AEAGLPDGVLNVVTGLGADVGAALAAHPGVDLVTFTGSTAVGRKVMAAAA
MUL_2789|M.ulcerans_Agy99 AEAGLPDGVLNVVTGLGADVGAALAAHPGVDLVTFTGSTAVGRKVMAAAA
MAV_2608|M.avium_104 AEAGFPPGVINVVTGWGPETGAALASHPGVDKVAFTGSTATGIHVGKAAI
TH_2957|M.thermoresistible__bu AEAGIPDGVINVVPGTG-DAGAALVAHPLVKKVTFTGGPDTARKILQSCA
MLBr_02573|M.leprae_Br4923 ARGGFPEDCFQTLLISANGVEAILR-DPRVAAATLTGSEPAGQAVGAIAG
..*: . ::.: . . : . : .:**. .. : .
Mb2883c|M.bovis_AF2122/97 AQVKRVTLELGGKSANIVFHDCDLERAAT-TAPAGVFDNAGQDCCARSRI
Rv2858c|M.tuberculosis_H37Rv AQVKRVTLELGGKSANIVFHDCDLERAAT-TAPAGVFDNAGQDCCARSRI
Mflv_4860|M.gilvum_PYR-GCK AHGHRTQLELGGKAPFVVFDDADLDAAIQ-GAVAGALINTGQDCTAATRA
Mvan_1572|M.vanbaalenii_PYR-1 AHGHRTQLELGGKAPFVVFDDADLDAAIQ-GAVAGALINTGQDCTAATRA
MSMEG_1665|M.smegmatis_MC2_155 VHGHRTQLELGGKAPFVVFSDADLDAAVQ-GAVAGALINTGQDCTAATRA
MAB_3328|M.abscessus_ATCC_1997 VHGHRTQLELGGKAPFVVFDDADMDAAIA-GAVAGAIINSGQDCTAATRA
MMAR_3556|M.marinum_M THGRRTQLELGGKAPFVVFDDADLDAAIQ-GAVAGALINSGQDCTAATRA
MUL_2789|M.ulcerans_Agy99 THGRRTQLELGGKAPFVVFDDADLDAAIQ-GAVAGAVINSGQDCTAATRA
MAV_2608|M.avium_104 ENMTRFSLELGGKSAQVVFADADLDAAAN-GVIAGVFAATGQTCLAGSRL
TH_2957|M.thermoresistible__bu EHIKPSVMELGGKSGNIIFEDANLDTACSVGTLMSVGMMSGQGCAFPTRM
MLBr_02573|M.leprae_Br4923 NEIKPTVLELGGSDPFIVMPSADLDAAVA-TAVTGRVQNNGQSCIAAKRF
. :****. ::: ..::: * . . ** * .*
Mb2883c|M.bovis_AF2122/97 LVQRSVYDRFMELLEPAVHSIVVGDPGSRATEMGPLVSRAHRDKVAGYVP
Rv2858c|M.tuberculosis_H37Rv LVQRSVYDRFMELLEPAVHSIVVGDPGSRATEMGPLVSRAHRDKVAGYVP
Mflv_4860|M.gilvum_PYR-GCK IVAEGLYDDFVAGVAEVMSKVVIGDPADPDTDLGPLISAVHREKVAGMVE
Mvan_1572|M.vanbaalenii_PYR-1 IVARDLYDDFVAGVAEVMSNIVVGDPGDPDTDLGPLITLAHRAKVAGMVE
MSMEG_1665|M.smegmatis_MC2_155 IVARELYDDFVAGVAEVMGKIVVGDPHDPDTDLGPLISYAHRDKVAGMVA
MAB_3328|M.abscessus_ATCC_1997 LVARDLYDDFVAGVGEVMSKVVVGDPLDPDTDIGPLISAAHRAKVSSIVD
MMAR_3556|M.marinum_M IVARELYHDFVAGVADVMSTVVIGDPMDPDTDIGPLISLAHRDRVADIVA
MUL_2789|M.ulcerans_Agy99 IVARELCHDFVAGVADVMSTVVIGDPMDPDTDIGPLISLAHRDRVADILA
MAV_2608|M.avium_104 LVDRSVADALVEKIVARAATIKLGDPKDPTTEMGPVSNLPQYEKVLSHFA
TH_2957|M.thermoresistible__bu IVARPLYDEVLTRVSALAPMVKVGDPLEPDTVAGPVVNEAALQRILGMIE
MLBr_02573|M.leprae_Br4923 IAHADIYDEFVEKFVARMETLRVGDPTDPDTDVGPLATESGRAQVEQQVD
:. : . .: . : :*** . * **: . :: .
Mb2883c|M.bovis_AF2122/97 ----DDAPVAFRGT--APAGR-GFWFPPTVLTPK-RGDRTVTDEIFGPVV
Rv2858c|M.tuberculosis_H37Rv ----DDAPVAFRGT--APAGR-GFWFPPTVLTPK-RGDRTVTDEIFGPVV
Mflv_4860|M.gilvum_PYR-GCK RAPGEGGRVVTGGV--APDGP-GSFYRPTLIADVAESSEVWRDEIFGPVL
Mvan_1572|M.vanbaalenii_PYR-1 RAPGEGGRIVTGGV--APDGP-GSFYRPTLIADVSEQSEVWRDEIFGPVL
MSMEG_1665|M.smegmatis_MC2_155 RAPQQGGRVVTGGE--APDLP-GAFYRPTLIADVDESSEVYRDEIFGPVL
MAB_3328|M.abscessus_ATCC_1997 RAPAQGGRIVTGAT--APDLP-GSFYRPTLIADVAETSEVYRDEIFGPVL
MMAR_3556|M.marinum_M KAPAQGARIVVGGR--IPDLA-GAFYLPTLLADVGEQSQAYREEIFGPVL
MUL_2789|M.ulcerans_Agy99 KAPAQGARIVVGGR--IPDLA-GAFYLPTLLADVGEQSQAYREEIFGPVL
MAV_2608|M.avium_104 SAREQGATVAYGGE-PASDLG-GFFVKPTVLTGVDPSMRAVAEEIFGPVL
TH_2957|M.thermoresistible__bu RAKQDGARLLTGGERLGGDLADEYFVAPTVFADVDPYSELAQKEVFGPVL
MLBr_02573|M.leprae_Br4923 AAAAAGAVIRCGGK--RPDRP-GWFYPPTVITDITKDMALYTDEVFGPVA
.. : . : **::: .*:****
Mb2883c|M.bovis_AF2122/97 VVLTFDDEADAISLANDTAYGLSGSIWTDDLSRALRVARAVESGNLSVNS
Rv2858c|M.tuberculosis_H37Rv VVLTFDDEADAISLANDTAYGLSGSIWTDDLSRALRVARAVESGNLSVNS
Mflv_4860|M.gilvum_PYR-GCK TVRSFTDDDDAIRQANDTKYGLAASAWTRDVYRAQRASREIHAGCVWIND
Mvan_1572|M.vanbaalenii_PYR-1 AVRSFTDDDDALRQANDTAYGLAASAWTRDVYRAQRASREIHAGCVWIND
MSMEG_1665|M.smegmatis_MC2_155 TVRSFTDDDDALRQANDTEYGLAASAWTRDVYRAQRASREINAGCVWIND
MAB_3328|M.abscessus_ATCC_1997 TVRPFTDDDDALRQANDTAYGLAASAWTRDVYRAQRASREIRAGCVWIND
MMAR_3556|M.marinum_M TVRAHDGDDDALRQANDTDYGLAASAWTRDVYRAQRASREISAGCVWIND
MUL_2789|M.ulcerans_Agy99 TVRAHDGDDDALRQANDTDYGLAASAWTRDVYRAQRASREISAGCVWIND
MAV_2608|M.avium_104 AVMTFEDEDEAVAAANGTEFGLAAAVWTKDVHRAHRVAAKLRAGTVWVNA
TH_2957|M.thermoresistible__bu SIIPFDTEEQAVEIANSTPYGLSGYVFTNDLKRAHRVAEQLETGEVLING
MLBr_02573|M.leprae_Br4923 SVYCATNIDDAIEIANATPFGLGSNAWTQNESEQRHFIDNIVAGQVFING
: :*: ** * :**.. :* : . : : :* : :*
Mb2883c|M.bovis_AF2122/97 HSSVRFNTPFGGFKQSGVGRELGPDAPLQFTETKNVFIAVGEEM------
Rv2858c|M.tuberculosis_H37Rv HSSVRFNTPFGGFKQSGVGRELGPDAPLQFTETKNVFIAVGEEM------
Mflv_4860|M.gilvum_PYR-GCK HIPIISEMPHGGFGASGFGKDMSQYSLEEYLSIKHVMSDITSAVDKDWHR
Mvan_1572|M.vanbaalenii_PYR-1 HIPIISEMPHGGFGASGFGKDMSQYSLEEYLSIKHVMSDITGVADKTWHR
MSMEG_1665|M.smegmatis_MC2_155 HIPIISEMPHGGVGASGFGKDMSDYSFEEYLTIKHVMSDITGVAEKEWHR
MAB_3328|M.abscessus_ATCC_1997 HIPIISEMPHGGFGASGFGKDMSDYSLEEYLTVKHVMSDITGTADKDWHR
MMAR_3556|M.marinum_M HIPIVSEMPHGGVAASGFGKDMSEYSFEEYLTIKHVMSDITGVAEKPWHR
MUL_2789|M.ulcerans_Agy99 HIPIVSEMPHGGVAASGFGNDMSEYSFDEYLTIKHVMSDITGVAEKPWHR
MAV_2608|M.avium_104 YRVVAPHVPFGGIGYSGIGRENGIDAVKDFTETKAVWVELSGETRDPFTL
TH_2957|M.thermoresistible__bu AAPLTVHRPFGGFGISGIGKEGGREGLAEFLRTKSVSIA-----------
MLBr_02573|M.leprae_Br4923 MTVSYPELPFGGIKRSGYGRELSTHGIREFCNIKTVWIA-----------
. *.**. ** *.: . . :: * *
Mb2883c|M.bovis_AF2122/97 -------
Rv2858c|M.tuberculosis_H37Rv -------
Mflv_4860|M.gilvum_PYR-GCK TIFTKR-
Mvan_1572|M.vanbaalenii_PYR-1 TIFTKR-
MSMEG_1665|M.smegmatis_MC2_155 TIFNKR-
MAB_3328|M.abscessus_ATCC_1997 TIFAKR-
MMAR_3556|M.marinum_M TIFSTRI
MUL_2789|M.ulcerans_Agy99 TIFSTRI
MAV_2608|M.avium_104 G------
TH_2957|M.thermoresistible__bu -------
MLBr_02573|M.leprae_Br4923 -------