For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MIQAVDRALRILTVLQGGRRMSLGEIASAIELAPSTVHGLVRTLLAHGMVQQELDSGRYRLGPATLRLGN VYLDTLELRSRVTVWAESLARRTGCAVRTAVLLFDEVVVVAHEPRPDGTRQMPEVGIVIPAHASALGKAM LAFQDDYRPEAPLRSMTGETVTDPEVLAAQLAQVRATGMATEAEEAVLGECSSAAAVFDATGEVAGAIAL VVPAARWPLAPDAVDALRDTARAVSRELGATVWPARRV
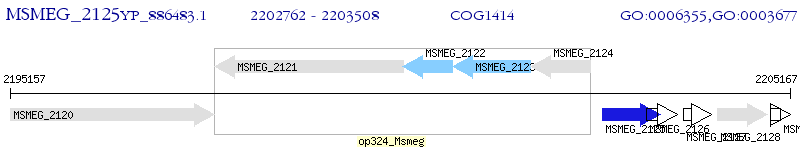
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2125 | - | - | 100% (248) | glycerol operon regulatory protein |
| M. smegmatis MC2 155 | MSMEG_6757 | - | 3e-36 | 43.32% (247) | glycerol operon regulatory protein |
| M. smegmatis MC2 155 | MSMEG_6118 | - | 2e-19 | 32.53% (249) | IclR-family protein transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1748 | - | 2e-16 | 30.94% (223) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_3087 | - | 4e-21 | 32.66% (248) | transcriptional regulator, TrmB |
| M. tuberculosis H37Rv | Rv1719 | - | 2e-16 | 30.94% (223) | transcriptional regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4493 | - | 1e-14 | 30.04% (223) | IclR family transcriptional regulator |
| M. marinum M | MMAR_4423 | - | 4e-23 | 34.01% (247) | transcriptional regulatory protein |
| M. avium 104 | MAV_3024 | - | 2e-17 | 31.06% (235) | transcriptional regulator, IclR family protein |
| M. thermoresistible (build 8) | TH_3647 | - | 6e-16 | 29.72% (249) | PUTATIVE putative transcriptional regulator, IclR family |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1018 | - | 4e-17 | 30.40% (227) | regulatory proteins, IclR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1748|M.bovis_AF2122/97 --------------MSAEEQDTRSGGIQVIARAAELLRVLQAHPG-GLSQ
Rv1719|M.tuberculosis_H37Rv --------------MSAEEQDTRSGGIQVIARAAELLRVLQAHPG-GLSQ
MAB_4493|M.abscessus_ATCC_1997 --------------------------MSVLTRAAQILDALAESNS-GLSV
Mflv_3087|M.gilvum_PYR-GCK --------MAQHDKTSTTSPTDAPAAVQSVDRALQVLEIVAAHG--QAGV
MMAR_4423|M.marinum_M MSSGISSMIMGTGNDGGASESDPAPTVQSVDRALLVLELLADLG--KAGV
MSMEG_2125|M.smegmatis_MC2_155 -------------------------MIQAVDRALRILTVLQGGR--RMSL
MAV_3024|M.avium_104 ------------------MPAPAETPTAVIDRISLVLDAFDGPG--RLTL
TH_3647|M.thermoresistible__bu -----------VRKRARGAIERPTYLLESVDNALRLLQMLRDGG--AVRL
Mvan_1018|M.vanbaalenii_PYR-1 ----------------MTEPDISVRKVKSAVRTVELLEYLAARPDDPARL
. :* .
Mb1748|M.bovis_AF2122/97 AEIGERVGMARSTVSRILNALEDEGLVASRGARGPYRLGPEITRMATTV-
Rv1719|M.tuberculosis_H37Rv AEIGERVGMARSTVSRILNALEDEGLVASRGARGPYRLGPEITRMATTV-
MAB_4493|M.abscessus_ATCC_1997 RELAEATGLPKSSVHRIVQELTVSQYVMAGGRGGSYQLGPGLLKLGLNN-
Mflv_3087|M.gilvum_PYR-GCK TEIAAELGVHKSTVSRLVAVLEARGYVEQVSERGKYRLGFAIPRLAHVAG
MMAR_4423|M.marinum_M TEVADELGVHKSTASRLITALEARGYVEQLSDRGKYQLGIGVSRLARARS
MSMEG_2125|M.smegmatis_MC2_155 GEIASAIELAPSTVHGLVRTLLAHGMVQQELDSGRYRLGPATLRLGNVYL
MAV_3024|M.avium_104 AQIVRRTGLPRSSAHRMLERLVQLRWLRRSGR--DYELGMRLVELGSLAV
TH_3647|M.thermoresistible__bu TEAAEELGVAPSTAHRLLAMLVYRGFAVQDEKR-TYLPGPALRAGPAERG
Mvan_1018|M.vanbaalenii_PYR-1 REISDALDMPRSSAHALLRTLVCQGWVRSDPTGTLYGIGIRALLVGTSYL
: : *:. :: * * *
Mb1748|M.bovis_AF2122/97 -RLGVVTEMHPFLTELSRELDETVDLSILDGDRADVVDQVVPPQRLRAVS
Rv1719|M.tuberculosis_H37Rv -RLGVVTEMHPFLTELSRELDETVDLSILDGDRADVVDQVVPPQRLRAVS
MAB_4493|M.abscessus_ATCC_1997 -RRQFVAPMRKAIATVAREVNENIDLAMLSGGELVIIEQIATVRNLPTVT
Mflv_3087|M.gilvum_PYR-GCK GQNDLVKLGQAACDMLSAGVGETANLAVLDADRIVNIVEAIGPSEITLRT
MMAR_4423|M.marinum_M GHSDLVKLSQDACDKLAADLGETANLAILDENRAVNIVEAVSTAEIALQT
MSMEG_2125|M.smegmatis_MC2_155 DTLELRSRVTVWAESLARRTGCAVRTAVLLFDEVVVVAHEPRPDGTRQMP
MAV_3024|M.avium_104 HQDRLVRAATPLLVELHRATGLVVHLAVLDGPDVVYLEKVGDRLNSVIPS
TH_3647|M.thermoresistible__bu WTREFTDVCLPHLEALSTVCGETVNLVIRVGTGVRFLTSVESTAMLRVGD
Mvan_1018|M.vanbaalenii_PYR-1 DSDPYLPLITPFLEDLRAEQDETFHLGRLDGTDIVYLATRESTQYTRTSS
: . :
Mb1748|M.bovis_AF2122/97 AVGESFPLYCCANGKALLAALPPERQARALPSRLAPLTANTITDRAALRD
Rv1719|M.tuberculosis_H37Rv AVGESFPLYCCANGKALLAALPPERQARALPSRLAPLTANTITDRAALRD
MAB_4493|M.abscessus_ATCC_1997 VVGRTFALHASSIGKALLSQLPPDQVRNMLPADLEVFTTNTITDVTALLR
Mflv_3087|M.gilvum_PYR-GCK WVGQSCPAYATSSGKVLLAGLDPAELPARIRPPLTEFTDSTLCGLAELQD
MMAR_4423|M.marinum_M WVGQSCPVHATSSGKVLLAGLDADEVRQRVGATLDSFTENTIVTIDALEA
MSMEG_2125|M.smegmatis_MC2_155 EVGIVIPAHASALGKAMLAFQDDYR----PEAPLRSMTGETVTDPEVLAA
MAV_3024|M.avium_104 RVGGRQPAHCTAVGKAILAYRDEATDLDLGARP----TKYSISSAPQLAA
TH_3647|M.thermoresistible__bu RRGQVLPAERTAGGRALLAELDDEVVER-------LYADRSAGQRAELLT
Mvan_1018|M.vanbaalenii_PYR-1 RVGRRLPAYATSLGKALLAERFGTERDEHVPAALKSLTPHTITDRTELDH
* . : *:.:*: : : *
Mb1748|M.bovis_AF2122/97 ELNRIRVDGVAYDREEQTEGICAVGAVLRG-VSVELVAVSVPVPAQRFYG
Rv1719|M.tuberculosis_H37Rv ELNRIRVDGVAYDREEQTEGICAVGAVLRG-VSVELVAVSVPVPAQRFYG
MAB_4493|M.abscessus_ATCC_1997 ELDDVRSTGVAFDHEEHDVGISSVAVPLPH-PAAIPQAMAIVAPTPRFAQ
Mflv_3087|M.gilvum_PYR-GCK ELAAVRERGWACVREELEVGLNAVAAPVYDVHAQVIAALSVSGPSYRLAV
MMAR_4423|M.marinum_M QLAATRAAGWASAREELEIGLNAVAAPVYDSTGRVAAALSVSGPAYRLPA
MSMEG_2125|M.smegmatis_MC2_155 QLAQVRATGMATEAEEAVLGECSSAAAVFDATGEVAGAIALVVPAARWPL
MAV_3024|M.avium_104 ELAKVRAHGVAFEREESLLGFGCVAAPIGG-PGEAAAAVSVCGPLQRMAL
TH_3647|M.thermoresistible__bu ELAAVREVGFGLNVEHTEEGVAAFGAVVRNRLGRAVGAVTVAVPIVRYQR
Mvan_1018|M.vanbaalenii_PYR-1 ALDEVRIRGYACDDEENTTGLRCFAVPLRY-CRPAQDAISASVPVERLDP
* * * . *. * . .. : *:: * *
Mb1748|M.bovis_AF2122/97 R-EAELAGALLAWVSKVDAWFNGTEDRK----------------------
Rv1719|M.tuberculosis_H37Rv R-EAELAGALLAWVSKVDAWFNGTEDRK----------------------
MAB_4493|M.abscessus_ATCC_1997 R-KTKYVAALDSLTKRANST------------------------------
Mflv_3087|M.gilvum_PYR-GCK D-RFDEVAKQTVAAAEQISRRLGWTTRN----------------------
MMAR_4423|M.marinum_M A-DFEEVAQQTIAAADAVSRGLGWTDRIS---------------------
MSMEG_2125|M.smegmatis_MC2_155 APDAVDALRDTARAVSRELGATVWPARRV---------------------
MAV_3024|M.avium_104 DQRLAAPVRMTAMGVWRNVEGGPQRVAPTLQRLRPLPPMRPQHPVPKPRP
TH_3647|M.thermoresistible__bu HARGDLVRQLFRTADEISVDLDGRFDSLEP--------------------
Mvan_1018|M.vanbaalenii_PYR-1 QRQREIIDALRAVGDKVSRVVRPLANGDKWFAS-----------------
Mb1748|M.bovis_AF2122/97 -----
Rv1719|M.tuberculosis_H37Rv -----
MAB_4493|M.abscessus_ATCC_1997 -----
Mflv_3087|M.gilvum_PYR-GCK -----
MMAR_4423|M.marinum_M -----
MSMEG_2125|M.smegmatis_MC2_155 -----
MAV_3024|M.avium_104 ALQYA
TH_3647|M.thermoresistible__bu -----
Mvan_1018|M.vanbaalenii_PYR-1 -----