For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TVAGQAHPPTRSGAAETRRAIWNTIRGSSGNLVEWYDVYVYTVFATYFEKQFFAEGDENSTVYVYAIFAV TFLTRPIGSWFFGRFADRRGRRAALTFSVSLMALCSLVIAIVPSREVIGAAAPVILILCRLVQGFATGGE YGTSATYMSEAATRERRGFFSSFQYVTLVGGHVLAQFTLLIILTAFTTEQVHDFGWRIGFAIGGVAAIVV FWLRRTMDESLSEEQLEAIKTGKDTSSGSLGELLTRYPKALLMCFLITLGGTIAFYTYSVNAPAIVKTTY KDQAMTATWINLFGLIFLMLLQPVGGLISDKVGRKPLLLWFGFGGVIYTYILITYLPQTHSAIMSFLLVA GGYIILTGYTSINAVVKAELFPSHIRALGVGVGYALANSLVGGTAPVIYQALKERDQVPLFIGYVTVCIA ISLVVYLFYLKNKSETYLDREKGAAFVR*
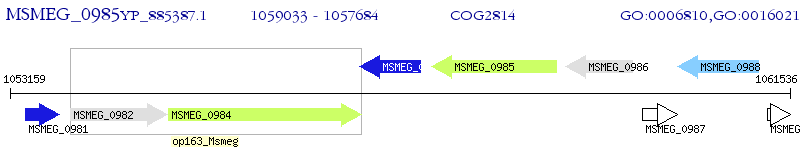
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0985 | - | - | 100% (449) | sugar transporter family protein |
| M. smegmatis MC2 155 | MSMEG_3990 | - | 2e-60 | 34.20% (421) | putative transporter |
| M. smegmatis MC2 155 | MSMEG_2952 | - | 3e-60 | 33.41% (422) | transport gene |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3503c | kgtP | 0.0 | 80.87% (439) | dicarboxylic acid transport integral membrane protein KgtP |
| M. gilvum PYR-GCK | Mflv_2381 | - | 5e-55 | 30.59% (425) | general substrate transporter |
| M. tuberculosis H37Rv | Rv3476c | kgtP | 0.0 | 80.87% (439) | dicarboxylic acid transport integral membrane protein KgtP |
| M. leprae Br4923 | MLBr_01562 | - | 4e-07 | 20.33% (359) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | MAB_3808c | - | 0.0 | 84.88% (443) | dicarboxylic acid transport integral membrane protein KgtP |
| M. marinum M | MMAR_4238 | - | 1e-29 | 25.73% (377) | integral membrane transport protein |
| M. avium 104 | MAV_4059 | - | 2e-32 | 27.32% (399) | monoxygenase |
| M. thermoresistible (build 8) | TH_3400 | - | 3e-53 | 31.17% (369) | PUTATIVE General substrate transporter |
| M. ulcerans Agy99 | MUL_0949 | - | 6e-30 | 25.73% (377) | integral membrane transport protein |
| M. vanbaalenii PYR-1 | Mvan_4266 | - | 5e-57 | 33.09% (417) | general substrate transporter |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0985|M.smegmatis_MC2_155 -------------MTVAGQAHPPTR-----SGAAETRRAIWNTIRGSS--
MAB_3808c|M.abscessus_ATCC_199 -------------MAVS-AADTSTP-----SGRAQTRRAIWNTIRGSS--
Mb3503c|M.bovis_AF2122/97 -------------MTVS--IAPPSR-----PSQAETRRAIWNTIRGSS--
Rv3476c|M.tuberculosis_H37Rv -------------MTVS--IAPPSR-----PSQAETRRAIWNTIRGSS--
MMAR_4238|M.marinum_M -----------------------------------MSRVALACVVG----
MUL_0949|M.ulcerans_Agy99 -----------------------------------MSRVALACVVG----
MAV_4059|M.avium_104 -------------MTLTSPAAASAG---DLPARPGLRTVAAGSMIG----
Mflv_2381|M.gilvum_PYR-GCK -------------MTVDEDQRRPS-----GAEPAAVRQAVRGAAIG----
Mvan_4266|M.vanbaalenii_PYR-1 -------------MTVKET--RP-------AAPGAVSRAVRGAAIG----
TH_3400|M.thermoresistible__bu -------------LTTERTDPPQDPSVEQEQPPGVLRKAIAASAVG----
MLBr_01562|M.leprae_Br4923 MVTLYDHERAVHNWTAARSDRPAPSRLTQQVEPASERTSKYPTLLPSGRF
MSMEG_0985|M.smegmatis_MC2_155 ------GNLVEWYDVYVYTVFATYFEKQFFAE--GDENSTVYVYAIFAVT
MAB_3808c|M.abscessus_ATCC_199 ------GNLVEWYDVYVYTVFAMYFEKQFFDD--SDQNSTVYVYAIFAVT
Mb3503c|M.bovis_AF2122/97 ------GNLVEWYDVYVYTVFATYFEDQFFDR--ADRNSTVYVYAIFAVT
Rv3476c|M.tuberculosis_H37Rv ------GNLVEWYDVYVYTVFATYFEDQFFDR--ADRNSTVYVYAIFAVT
MMAR_4238|M.marinum_M -------STVEYYDFCIYGTAAALVFPAVFYPGLSPALATIASMGTFATA
MUL_0949|M.ulcerans_Agy99 -------STVEYYDFCIYGTAAALVFPAVFYPGLSPALATIASMGTFATA
MAV_4059|M.avium_104 -------TTIEWYDFYLYATASALVFKPLFFPNISPSAGTLASFATYAAG
Mflv_2381|M.gilvum_PYR-GCK -------NTVEWFDFAIYGFLATYIA-DKFFPSGDETAALLSTFAVFAAA
Mvan_4266|M.vanbaalenii_PYR-1 -------NAVEWFDFAIYGFLATYIA-EKFFPPGDETAALLNTFAIFAAA
TH_3400|M.thermoresistible__bu -------NATEWFDYGLYAYGVTYIS-VAIFP-GDGATATLLALMTFAVS
MLBr_01562|M.leprae_Br4923 PSGRFIAAVIVIGGMQLLATMDSTVAIVALPKIQNELSLSDAGRSWGITA
. : . : .
MSMEG_0985|M.smegmatis_MC2_155 FLTRPIGSWFFG-RFADRRGRRAALTFSVSLMALCSLVIAIVPSREVIGA
MAB_3808c|M.abscessus_ATCC_199 FITRPLGSWFFG-RFADRNGRRAALTLSVSLMAACSMVIALVPSRGSIGM
Mb3503c|M.bovis_AF2122/97 FVTRPVGSWFFG-RFADRRGRRAALTFSVSLMAACSLIVALVPSRSSIGV
Rv3476c|M.tuberculosis_H37Rv FVTRPVGSWFLG-RFADRRGRRAALTFSVSLMAACSLIVALVPSRSSIGV
MMAR_4238|M.marinum_M FLSRPLGAAVFG-HFGDRLGRKKTLVATLLIMALSTVTVGVVPSTATIGV
MUL_0949|M.ulcerans_Agy99 FLSRPLGAAVFG-HFGDRLGRKKTLVATLLIMALSTVTVGVVPSTATIGV
MAV_4059|M.avium_104 FGARPLGAVLSG-HFGDRLGRKTVLVAALLVMGLVTTAIGALPTYAEAGL
Mflv_2381|M.gilvum_PYR-GCK FFMRPLGGFFFG-PLGDRIGRQRVLAIVILLMSASTLAIGLVPSYDSIGV
Mvan_4266|M.vanbaalenii_PYR-1 FFMRPLGGFFFG-PLGDRIGRQRVLALVILLMSASTLAIGLVPGYDSIGV
TH_3400|M.thermoresistible__bu FLVRPLGGFVWG-PLGDRLGRRQVLAITILLMAGATFCVGLVPSYDSIGL
MLBr_01562|M.leprae_Br4923 YVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCAVA--WDEVTL
: * . * :.* **: .: : :: : .
MSMEG_0985|M.smegmatis_MC2_155 AAPVILILCRLVQGFATGGEYGTSATYMSEAATRERRGFFSSFQYVTLVG
MAB_3808c|M.abscessus_ATCC_199 AAPIVLILCRLVQGFATGGEYGTSATYMSEAATRERRGFFSSFQYVTLVG
Mb3503c|M.bovis_AF2122/97 AAPILLILCRLVQGFATGGEYGTSVTYMSEAATRERRGYFSSFQYVTLVG
Rv3476c|M.tuberculosis_H37Rv AAPILLILCRLVQGFATGGEYGTSATYMSEAATRERRGYFSSFQYVTLVG
MMAR_4238|M.marinum_M SAPLILVALRLLQGFAVGGEWAGSALLSIESAPAHKRGYYGMFTVLGGGV
MUL_0949|M.ulcerans_Agy99 SAPLILVALRLLQGFAVGGEWAGSALLSIESAPAHKRGYYGMFTVLGGGV
MAV_4059|M.avium_104 AAPALLASLRVVQGLAVGAEWGGAAVLSVEHAPPGRRGLFGSFTQLGSPA
Mflv_2381|M.gilvum_PYR-GCK LAPILLLVLRCLQGFSAGGEYGSGACFLAEYAPNRHRGFVVSFLVWSVVV
Mvan_4266|M.vanbaalenii_PYR-1 LAPILLLLLRCLQGFSAGGEYGSGACFLAEYAPDRHRGFVVSFLVWSVVV
TH_3400|M.thermoresistible__bu WAPVLMVILRMIQGFSTGGEYGGAATFMAEYAPSRRRGLLGSFLEFGTLA
MLBr_01562|M.leprae_Br4923 VIARLLQGIGSAIASPTG-LALVATTFSKGSARNTATAVFAAMTAIGSVM
. :: . ..* .. * . :
MSMEG_0985|M.smegmatis_MC2_155 GHVLAQFTLLIILTAFT--TEQVHDFGWRIGFAIGGVAAIVVFWLRRTMD
MAB_3808c|M.abscessus_ATCC_199 GHVLAQFTLLIILTVFD--TAQVHEFGWRIAFATGGVAAIVVYWLRRTMD
Mb3503c|M.bovis_AF2122/97 GHVLAQFTLLVILAVFT--REQVHEFGWRIGFAVGGGAAIVVFWLRRTMD
Rv3476c|M.tuberculosis_H37Rv GHVLAQFTLLVILAVFT--REQVHEFGWRIGFAVGGGAAIVVFWLRRTMD
MMAR_4238|M.marinum_M ALVVSGLTFLGVNYTIGESSSAFLQWGWRVPFLLSALLIAFALYVRLEIN
MUL_0949|M.ulcerans_Agy99 ALVVSGLTFLGVNYTIGESSSALLQWGWRVPFLLSALLIAFALYVRLEIN
MAV_4059|M.avium_104 GMLLATSVFFGVRKATG--PAAFLGFGWRIPFLLSIFLVAVGLFVRLRLT
Mflv_2381|M.gilvum_PYR-GCK GFLAGAITVTALETWLS--EDAMNSYGWRIPFMIAGLLGAVGLYIRLKLG
Mvan_4266|M.vanbaalenii_PYR-1 GFLLGSVTVTALEATLS--ESAMNSFGWRIPFLIAGALGAVGLYIRLRLG
TH_3400|M.thermoresistible__bu GFSLGALMMLGVSLMLS--DDQMNTWGWRVPFLIAAPLGLIGLYLRSRLD
MLBr_01562|M.leprae_Br4923 GLVVG---------------GALTEVSWRLAFLVNVPIGLVMIYLARTAL
. . . .**: * . ::
MSMEG_0985|M.smegmatis_MC2_155 ESLSEEQLEAIKTGKDTS---SGSLGELLTRYPKALLMCFLITLGGTIAF
MAB_3808c|M.abscessus_ATCC_199 ESLSEEQLAAIKAGADTS---SGSMRELLTRYRKPLLLCFLITMGGTLAF
Mb3503c|M.bovis_AF2122/97 ESLSQERLTAIKAGRDHD---SGSLRELATHYWKPLLLCFLVTLGGTVAF
Rv3476c|M.tuberculosis_H37Rv ESLSQERLTAIKAGRDHD---SGSLRELATHYWKPLLLCFLVTLGGTVAF
MMAR_4238|M.marinum_M ETPVFARAMAQADDQESTAAARAPLAAVLCRQRREIVLAAGAMLAAFGCL
MUL_0949|M.ulcerans_Agy99 ETPVFARAMAQADDQESTAAARAPLAAVLCRQRREIVLAAGAMLAAFGCL
MAV_4059|M.avium_104 DAEVFDRLRSRDE------LARLPIVQVLRTDARNVVITTGLRLSQIGLF
Mflv_2381|M.gilvum_PYR-GCK DTPEFRSLRESGEVSQ------SPLREAVTTSWRPILQIIGLVVIHNVGF
Mvan_4266|M.vanbaalenii_PYR-1 DTPEFEALRESGEVST------TPLKEAVTTSWRPILQIIGLVVVHNVGF
TH_3400|M.thermoresistible__bu DTPIFRELQESDREEKS---IWTEFKDLLVEYRGQILRLGGLVVALNVVN
MLBr_01562|M.leprae_Br4923 HETHKERMKLDAAGAILATLACTAAVFAFSMGPEKGWISLTTIGSGLVAL
.
MSMEG_0985|M.smegmatis_MC2_155 YTYSVNAPAIVKTTYKDQAMTATWINLFGLIFLMLLQPVGG--LISDKVG
MAB_3808c|M.abscessus_ATCC_199 YTYSVNAPAIVKAAYKDQAMTATWINLAGLIFLMLLQPVGG--IISDKVG
Mb3503c|M.bovis_AF2122/97 YTYSVNAPAIVKSVYGSQAMTATWINLVGLILLMMLQPIGG--MISDKIG
Rv3476c|M.tuberculosis_H37Rv YTYSVNAPAIVKSVYGSQAMTATWINLVGLILLMMLQPIGG--MISDKIG
MMAR_4238|M.marinum_M YLASTFLPSYAQLHLGYSRNLVLLIGVLGGLVCIGFVALS--AILCDRFG
MUL_0949|M.ulcerans_Agy99 YLASTFLPSYAQLHLGYSRNLVLLIGVLGGLVCIGFVALS--AILCDRFG
MAV_4059|M.avium_104 VLLTTYSLSYLQDSFGKGSGVGLVAVLIS--SALGFLSTPGWALLSDRVG
Mflv_2381|M.gilvum_PYR-GCK YIVYTFLPSYFTETLDFSKVDAFWSITVASLVGIVLIPPLG--ALSDRIG
Mvan_4266|M.vanbaalenii_PYR-1 YVVYTFLPSYFTRTLEFSKIDAFYSITVASLVGIVLIPPLG--ALSDRVG
TH_3400|M.thermoresistible__bu YTLLSYMPTYLQTSIGLSTDTALVVPIIGMLSMMVFLPFAG--LLSDRFG
MLBr_01562|M.leprae_Br4923 VAAIAFAVVERTAENPVVPFDLFRDRNRLVTFIAIFLAGGVIFSLTVSIG
: . : .*
MSMEG_0985|M.smegmatis_MC2_155 RKPLLLWFGFGGVIYTYILITYLPQTHSAIMSFLLVAGGYIILTGYTSIN
MAB_3808c|M.abscessus_ATCC_199 RKPLLLFFGFGGVVYTYVLITYLPQVHAPIVSFLLVAVSYVLLTGYTSIN
Mb3503c|M.bovis_AF2122/97 RKPLLLWFGVGGLIYTYVLVTYLPETRSPTMSFLLVAVGYVILTGYCSIN
Rv3476c|M.tuberculosis_H37Rv RKPLLLWFGVGGLIYTYVLVTYLPETRSPTMSFLLVAVGYVILTGYCSIN
MMAR_4238|M.marinum_M RRRVMLVGWAVGLFWSPLVMPLIGSGDTVGFTVALLG-LYAVAASGFGPA
MUL_0949|M.ulcerans_Agy99 RRRVMLVGWAVGLFWSPLVMPLIGSGDTVGFTVALLG-LYAVAASGFGPA
MAV_4059|M.avium_104 RRPPYLFGALASVVALVLFFVAAGTGSAVLVVVAIVFGVNVVHDAMYGPQ
Mflv_2381|M.gilvum_PYR-GCK RKSLLIAGAVAFAVFAYPLFLLLNTGSLAAAIAAHAA-LAAIESVFVSAS
Mvan_4266|M.vanbaalenii_PYR-1 RKPLLLAGSIGFAVFAYPLFLLLNTGSLAAAIAAHAA-LAAIEAVFVCAS
TH_3400|M.thermoresistible__bu RKPMWWISLGGLLALVVPMFMLMST-SVPGAIIGFAV-LGLLYVPQLATI
MLBr_01562|M.leprae_Br4923 LYIQDILGYSALRAGIGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGW
.
MSMEG_0985|M.smegmatis_MC2_155 AVVKAELFPSHIRALGVGVGYALANSLVGG-------TAPVIYQALKER-
MAB_3808c|M.abscessus_ATCC_199 ALVKSELFPSHVRALGVGVGYALANSVFGG-------TAPLIYQALKEH-
Mb3503c|M.bovis_AF2122/97 ALVKSELFPAHVRALGVGVGYALANSVFGG-------TAPLIYQALKER-
Rv3476c|M.tuberculosis_H37Rv ALVKSELFPAHVRALGVGVGYALANSVFGG-------TAPLIYQALKER-
MMAR_4238|M.marinum_M ASLIPELFATRYRYTGTALALNVAGIVGGA-------APPLIAGTLLAS-
MUL_0949|M.ulcerans_Agy99 ASLIPELFATRYRYTGTALALNVAGIVGGA-------APPLIAGTLLAS-
MAV_4059|M.avium_104 AAWFAELFDTRVRYSGSSLGYHIGAVLSGG-------FAPLIAASLLVAG
Mflv_2381|M.gilvum_PYR-GCK LAAGAELFATKVRSSGYSIGYNVSVAVFGG-------TAPYVATWLVSR-
Mvan_4266|M.vanbaalenii_PYR-1 LAAGAELFATRVRSSGYSIGYNVSVAIFGG-------TAPYVATWLVSR-
TH_3400|M.thermoresistible__bu SATFPAMFPTHVRYAGFAIAYNVSTSLFGG-------TAPAANDWLIQQ-
MLBr_01562|M.leprae_Br4923 LLMVAMLYGWAFMNRGVPYFPNLLAPIVVGGIGIGMAVVPLTLSAIAGVG
. :: * : : . * :
MSMEG_0985|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3808c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3503c|M.bovis_AF2122/97 --------------------------------------------------
Rv3476c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4238|M.marinum_M --------------------------------------------------
MUL_0949|M.ulcerans_Agy99 --------------------------------------------------
MAV_4059|M.avium_104 GGRPWLIVGYFAVLAAITVGAACAGPGDARGADRMSAARRIYLNAFDMAC
Mflv_2381|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4266|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3400|M.thermoresistible__bu --------------------------------------------------
MLBr_01562|M.leprae_Br4923 ----------------------------------------------FDRI
MSMEG_0985|M.smegmatis_MC2_155 ---DQVPLFIG-------------------------YVTVCIAISLVVYL
MAB_3808c|M.abscessus_ATCC_199 ---DQVPLFIG-------------------------YVTVCIAISLVVYL
Mb3503c|M.bovis_AF2122/97 ---DQVPMFIA-------------------------YVTACIAVSLIVYV
Rv3476c|M.tuberculosis_H37Rv ---DQVPMFIA-------------------------YVTACIAVSLIVYV
MMAR_4238|M.marinum_M YGSSAVGLMMT---------------------------ALVAASLLCTYL
MUL_0949|M.ulcerans_Agy99 YGSSAVGLMMT---------------------------ALVAASLLCTYL
MAV_4059|M.avium_104 VGHQSAGLWRHPDDQGHRYRELSYWTELARTLEAGGFDALFLADVLGVYD
Mflv_2381|M.gilvum_PYR-GCK TGNDIAPAYYL--------------------------IAAAIVSLLTVLT
Mvan_4266|M.vanbaalenii_PYR-1 TGNDIAPAFYV--------------------------IAAAVVTFLTVLT
TH_3400|M.thermoresistible__bu TGNNLMPAFYM--------------------------MAACLVGMLALVK
MLBr_01562|M.leprae_Br4923 GPVSAIALMLQSLG--------------------GPLVLAVIQAVITSRT
. :
MSMEG_0985|M.smegmatis_MC2_155 FYLK----------------------------NKSETYLDREKGAAFVR-
MAB_3808c|M.abscessus_ATCC_199 FFLK----------------------------NKSQTYLDREQGSAFNR-
Mb3503c|M.bovis_AF2122/97 FFIK----------------------------NKADTYLDREQGFAFYGH
Rv3476c|M.tuberculosis_H37Rv FFIK----------------------------NKADTYLDREQGFAFYGH
MMAR_4238|M.marinum_M LP------------------------------ETAGAPLTAA--------
MUL_0949|M.ulcerans_Agy99 LP------------------------------ETAGAPLTAA--------
MAV_4059|M.avium_104 IYGASRDAAVVDAAQFPVNEPSAAVSAMAAVTETLGFGITLSLTYEQPYA
Mflv_2381|M.gilvum_PYR-GCK MR------------------------------ETARKPLR----------
Mvan_4266|M.vanbaalenii_PYR-1 MR------------------------------ETARQPLR----------
TH_3400|M.thermoresistible__bu LP------------------------------ETARCPIGGTEIPGTEDA
MLBr_01562|M.leprae_Br4923 LYLG-----------------------------GTTGPVKFMNDAQLRAL
: :
MSMEG_0985|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3808c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3503c|M.bovis_AF2122/97 A-------------------------------------------------
Rv3476c|M.tuberculosis_H37Rv A-------------------------------------------------
MMAR_4238|M.marinum_M --------------------------------------------------
MUL_0949|M.ulcerans_Agy99 --------------------------------------------------
MAV_4059|M.avium_104 LARRLSTLDHLTGGRVAWNIVTSYLDSAARNLGLDAQIPHDQRYEIADEY
Mflv_2381|M.gilvum_PYR-GCK ------KLVHP---------------------------------------
Mvan_4266|M.vanbaalenii_PYR-1 ------KLVSD---------------------------------------
TH_3400|M.thermoresistible__bu PPPVEYDLVGNR--------------------------------------
MLBr_01562|M.leprae_Br4923 DHGYTYGLLWVAGAVVIVGVAALFIGYTPEQVAYAQEVKEAVDAGEL---
MSMEG_0985|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3808c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3503c|M.bovis_AF2122/97 --------------------------------------------------
Rv3476c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4238|M.marinum_M --------------------------------------------------
MUL_0949|M.ulcerans_Agy99 --------------------------------------------------
MAV_4059|M.avium_104 LEVCYKLWEGSWEAGAVLRDRQRGVFTDPAKVHDIEHKGRYFSVPGPFLC
Mflv_2381|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4266|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3400|M.thermoresistible__bu --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
MSMEG_0985|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3808c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3503c|M.bovis_AF2122/97 --------------------------------------------------
Rv3476c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4238|M.marinum_M --------------------------------------------------
MUL_0949|M.ulcerans_Agy99 --------------------------------------------------
MAV_4059|M.avium_104 EPSPQRTPVLFQAGASPRGIRFAAAHAEAVFVSGPTAQVVREPVRALRTA
Mflv_2381|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4266|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3400|M.thermoresistible__bu --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
MSMEG_0985|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3808c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3503c|M.bovis_AF2122/97 --------------------------------------------------
Rv3476c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4238|M.marinum_M --------------------------------------------------
MUL_0949|M.ulcerans_Agy99 --------------------------------------------------
MAV_4059|M.avium_104 AAQLGRDPRAVKVFTMVTPVVAETHDAAVAKLAEYRRFVSPRGALALFGG
Mflv_2381|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4266|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3400|M.thermoresistible__bu --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
MSMEG_0985|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3808c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3503c|M.bovis_AF2122/97 --------------------------------------------------
Rv3476c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4238|M.marinum_M --------------------------------------------------
MUL_0949|M.ulcerans_Agy99 --------------------------------------------------
MAV_4059|M.avium_104 WTGVDLAELDPDEPLTYVETDANRSALASFTTAGRAWTVAELAAEIGIGG
Mflv_2381|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4266|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3400|M.thermoresistible__bu --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
MSMEG_0985|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3808c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3503c|M.bovis_AF2122/97 --------------------------------------------------
Rv3476c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4238|M.marinum_M --------------------------------------------------
MUL_0949|M.ulcerans_Agy99 --------------------------------------------------
MAV_4059|M.avium_104 RGPVLVGSPTEVADELERWVDDADVDGFNVAYVTTPGTFVDFAKFVVPEL
Mflv_2381|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4266|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3400|M.thermoresistible__bu --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
MSMEG_0985|M.smegmatis_MC2_155 -----------------------------------------
MAB_3808c|M.abscessus_ATCC_199 -----------------------------------------
Mb3503c|M.bovis_AF2122/97 -----------------------------------------
Rv3476c|M.tuberculosis_H37Rv -----------------------------------------
MMAR_4238|M.marinum_M -----------------------------------------
MUL_0949|M.ulcerans_Agy99 -----------------------------------------
MAV_4059|M.avium_104 RRRGHVPEQVPGSTLRERLGAAGPLLGDDHPGAAYRLRRDR
Mflv_2381|M.gilvum_PYR-GCK -----------------------------------------
Mvan_4266|M.vanbaalenii_PYR-1 -----------------------------------------
TH_3400|M.thermoresistible__bu -----------------------------------------
MLBr_01562|M.leprae_Br4923 -----------------------------------------