For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TVDEDQRRPSGAEPAAVRQAVRGAAIGNTVEWFDFAIYGFLATYIADKFFPSGDETAALLSTFAVFAAAF FMRPLGGFFFGPLGDRIGRQRVLAIVILLMSASTLAIGLVPSYDSIGVLAPILLLVLRCLQGFSAGGEYG SGACFLAEYAPNRHRGFVVSFLVWSVVVGFLAGAITVTALETWLSEDAMNSYGWRIPFMIAGLLGAVGLY IRLKLGDTPEFRSLRESGEVSQSPLREAVTTSWRPILQIIGLVVIHNVGFYIVYTFLPSYFTETLDFSKV DAFWSITVASLVGIVLIPPLGALSDRIGRKSLLIAGAVAFAVFAYPLFLLLNTGSLAAAIAAHAALAAIE SVFVSASLAAGAELFATKVRSSGYSIGYNVSVAVFGGTAPYVATWLVSRTGNDIAPAYYLIAAAIVSLLT VLTMRETARKPLRKLVHP*
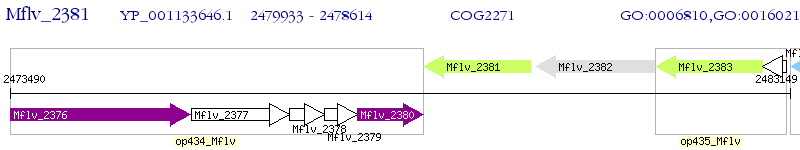
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2381 | - | - | 100% (439) | general substrate transporter |
| M. gilvum PYR-GCK | Mflv_1075 | - | e-169 | 67.78% (419) | general substrate transporter |
| M. gilvum PYR-GCK | Mflv_0953 | - | e-102 | 42.57% (444) | general substrate transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3503c | kgtP | 5e-58 | 32.86% (420) | dicarboxylic acid transport integral membrane protein KgtP |
| M. tuberculosis H37Rv | Rv3476c | kgtP | 8e-58 | 32.86% (420) | dicarboxylic acid transport integral membrane protein KgtP |
| M. leprae Br4923 | MLBr_00556 | - | 6e-05 | 30.68% (88) | putative integral membrane drug transport protein |
| M. abscessus ATCC 19977 | MAB_2263c | - | 3e-84 | 39.49% (428) | major facilitator transporter |
| M. marinum M | MMAR_4238 | - | 5e-56 | 33.09% (417) | integral membrane transport protein |
| M. avium 104 | MAV_4627 | - | 5e-64 | 33.95% (430) | major facilitator superfamily protein |
| M. smegmatis MC2 155 | MSMEG_4889 | - | 1e-165 | 67.63% (417) | integral membrane transport protein |
| M. thermoresistible (build 8) | TH_3722 | - | 1e-161 | 64.86% (424) | PUTATIVE General substrate transporter |
| M. ulcerans Agy99 | MUL_0949 | - | 2e-56 | 33.09% (417) | integral membrane transport protein |
| M. vanbaalenii PYR-1 | Mvan_4266 | - | 0.0 | 88.24% (425) | general substrate transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3503c|M.bovis_AF2122/97 ------------MTVSIAPPSRPSQAETRRAIWNTIRGS--SGNLVEWYD
Rv3476c|M.tuberculosis_H37Rv ------------MTVSIAPPSRPSQAETRRAIWNTIRGS--SGNLVEWYD
Mflv_2381|M.gilvum_PYR-GCK ----------MTVDEDQRRPSGAEPAAVR---QAVRGAA--IGNTVEWFD
Mvan_4266|M.vanbaalenii_PYR-1 ----------MTVKET--RP--AAPGAVS---RAVRGAA--IGNAVEWFD
MSMEG_4889|M.smegmatis_MC2_155 ------------------MAVTDRETAVR---RAVMGAS--IGNAVEWFD
TH_3722|M.thermoresistible__bu ---------VSENPSIVSAPPAPDAVVIR---KAVTGAS--IGNAVEWFD
MAB_2263c|M.abscessus_ATCC_199 -MSSAGPREGETEPPPVPRPDFQSPEGRRQLHRAIAASA--MGNATEWYD
MMAR_4238|M.marinum_M ------------------------------MSRVALACV--VGSTVEYYD
MUL_0949|M.ulcerans_Agy99 ------------------------------MSRVALACV--VGSTVEYYD
MAV_4627|M.avium_104 -MAATLPAPAPAGLADGHPLDESAEQQRRRLRIVVAASL--LGTTVEWYD
MLBr_00556|M.leprae_Br4923 MSTRAGRRVAISAGSLAVLLGALDTYVVVTIMRDIMHDVGIPVNQMQRIT
. :
Mb3503c|M.bovis_AF2122/97 VYVYTVFATYFEDQFFDR--ADRNSTVYVYAIFAVTFVTRPVGSWFFGRF
Rv3476c|M.tuberculosis_H37Rv VYVYTVFATYFEDQFFDR--ADRNSTVYVYAIFAVTFVTRPVGSWFLGRF
Mflv_2381|M.gilvum_PYR-GCK FAIYGFLATYIA-DKFFPSGDETAALLSTFAVFAAAFFMRPLGGFFFGPL
Mvan_4266|M.vanbaalenii_PYR-1 FAIYGFLATYIA-EKFFPPGDETAALLNTFAIFAAAFFMRPLGGFFFGPL
MSMEG_4889|M.smegmatis_MC2_155 FAIYGFLATSIA-ANFFPAGDETAALLNTFAIFAAAFFMRPLGGFVFGPL
TH_3722|M.thermoresistible__bu FAIYGFLATYIA-ANFFPAGDDTAALLNTFAIFAAAFVVRPLGGFFFGPL
MAB_2263c|M.abscessus_ATCC_199 YGVYAATTTFLT-QAFFPTLGTAFTMLG----YAISFLLRPLGGMVWGPL
MMAR_4238|M.marinum_M FCIYGTAAALVFPAVFYPGLSPALATIASMGTFATAFLSRPLGAAVFGHF
MUL_0949|M.ulcerans_Agy99 FCIYGTAAALVFPAVFYPGLSPALATIASMGTFATAFLSRPLGAAVFGHF
MAV_4627|M.avium_104 FFLYATAAGLVFNKVFFPNESSLVGTLLAFATFAVGFVMRPIGGLVFGHI
MLBr_00556|M.leprae_Br4923 WIVTMYLLGYIAAMPLLSRASDRFGRKLLLQVSLAGFAIGSVMTALAGQF
: . : * .: . * :
Mb3503c|M.bovis_AF2122/97 ADR-----------------RGRRAALTFSVSLMAACSLIVALVPSRSSI
Rv3476c|M.tuberculosis_H37Rv ADR-----------------RGRRAALTFSVSLMAACSLIVALVPSRSSI
Mflv_2381|M.gilvum_PYR-GCK GDR-----------------IGRQRVLAIVILLMSASTLAIGLVPSYDSI
Mvan_4266|M.vanbaalenii_PYR-1 GDR-----------------IGRQRVLALVILLMSASTLAIGLVPGYDSI
MSMEG_4889|M.smegmatis_MC2_155 GDR-----------------LGRQRVLAIVILLMSAATLGIGLLPTYSTI
TH_3722|M.thermoresistible__bu GDR-----------------IGRQRVLALVILLMSGATLTMGLLPTYESI
MAB_2263c|M.abscessus_ATCC_199 GDR-----------------LGRKSVMAITIVLMALSTTLIGILPSWATI
MMAR_4238|M.marinum_M GDR-----------------LGRKKTLVATLLIMALSTVTVGVVPSTATI
MUL_0949|M.ulcerans_Agy99 GDR-----------------LGRKKTLVATLLIMALSTVTVGVVPSTATI
MAV_4627|M.avium_104 GDR-----------------IGRKRSLALTMLIMGGATALIGVLPTAAQI
MLBr_00556|M.leprae_Br4923 GDFHMLIAGRTIQGVASGALLPITLALGADLWAQRNRAGVLGGIGAAQEL
.* : : : :. : :
Mb3503c|M.bovis_AF2122/97 G-VAAPILLILCRLVQG-----FATGGEYGTSVTYMSEAATRERRGYFSS
Rv3476c|M.tuberculosis_H37Rv G-VAAPILLILCRLVQG-----FATGGEYGTSATYMSEAATRERRGYFSS
Mflv_2381|M.gilvum_PYR-GCK G-VLAPILLLVLRCLQG-----FSAGGEYGSGACFLAEYAPNRHRGFVVS
Mvan_4266|M.vanbaalenii_PYR-1 G-VLAPILLLLLRCLQG-----FSAGGEYGSGACFLAEYAPDRHRGFVVS
MSMEG_4889|M.smegmatis_MC2_155 G-VVAPLLLLLLRCLQG-----FSAGGEYGGGAVYLAEYATQRRRGLTVT
TH_3722|M.thermoresistible__bu G-VLAPLLLLLLRCLQG-----FSAGGEYGGGAVYLAEFAPDRRRGLIVT
MAB_2263c|M.abscessus_ATCC_199 G-MAAPALLILLRVIQG-----FSTGGEYGGAATFMAEFAPDKKRGKYGS
MMAR_4238|M.marinum_M G-VSAPLILVALRLLQG-----FAVGGEWAGSALLSIESAPAHKRGYYGM
MUL_0949|M.ulcerans_Agy99 G-VSAPLILVALRLLQG-----FAVGGEWAGSALLSIESAPAHKRGYYGM
MAV_4627|M.avium_104 G-AWAPVLLLVLRVLQG-----FALGGEWGGAVLLAVEHSPGDRRGRYGA
MLBr_00556|M.leprae_Br4923 GSVLGPLYGIFIVWLFSDWRYVFWINIPLTAIAMLMIQVSLSAHD-RGDE
* .* : : . * . . : : :
Mb3503c|M.bovis_AF2122/97 FQYVTLVGGHVLAQFTLLVILAVFT--REQVHEFG-WRIGFAVGGGAAIV
Rv3476c|M.tuberculosis_H37Rv FQYVTLVGGHVLAQFTLLVILAVFT--REQVHEFG-WRIGFAVGGGAAIV
Mflv_2381|M.gilvum_PYR-GCK FLVWSVVVGFLAGAITVTALETWLS--EDAMNSYG-WRIPFMIAGLLGAV
Mvan_4266|M.vanbaalenii_PYR-1 FLVWSVVVGFLLGSVTVTALEATLS--ESAMNSFG-WRIPFLIAGALGAV
MSMEG_4889|M.smegmatis_MC2_155 FIAWSGVLGFLLGSITVTVLAALLP--PEAMDTYG-WRIPFLIAAPLGLV
TH_3722|M.thermoresistible__bu FMAWSGVLGFLLGSVTVTALQVALP--DQVMTDVG-WRIPFLLAGPLGLI
MAB_2263c|M.abscessus_ATCC_199 FLEFGTLGGFAVGAAFVLVLDATLS--DDAMHTWG-WRIPFLVALPMGLI
MMAR_4238|M.marinum_M FTVLGGGVALVVSGLTFLGVNYTIGESSSAFLQWG-WRVPFLLSALLIAF
MUL_0949|M.ulcerans_Agy99 FTVLGGGVALVVSGLTFLGVNYTIGESSSALLQWG-WRVPFLLSALLIAF
MAV_4627|M.avium_104 VPQVGLALGLALGTGIFAFLQIVLG--PARFLSYG-WRIGFLLSVLLVAI
MLBr_00556|M.leprae_Br4923 LEKVDVVGGVLLAIALGLVVIGLYNPQPDSKQVLPSYGVPVLVGGIVATV
. . . : : : . :. .
Mb3503c|M.bovis_AF2122/97 VFWLRRTMDES----LSQERLTAIKAG---RDHDSGSLRELAT--HYWKP
Rv3476c|M.tuberculosis_H37Rv VFWLRRTMDES----LSQERLTAIKAG---RDHDSGSLRELAT--HYWKP
Mflv_2381|M.gilvum_PYR-GCK GLYIRLKLGDT----PEFRSLRESG------EVSQSPLREAVT--TSWRP
Mvan_4266|M.vanbaalenii_PYR-1 GLYIRLRLGDT----PEFEALRESG------EVSTTPLKEAVT--TSWRP
MSMEG_4889|M.smegmatis_MC2_155 GLYIRLRLDDT----PEFEKLDAAD------RVAKSPLREALR--TARKP
TH_3722|M.thermoresistible__bu GLYIRLRLDDT----PAFTALSEQE------EVAQSPLREAVQ--TAWRS
MAB_2263c|M.abscessus_ATCC_199 GWYLRSRLDDP----PLFKEMQEQEPEGR--ETAFERLRDLVR--DYRRP
MMAR_4238|M.marinum_M ALYVRLEINET----PVFARAMAQADDQESTAAARAPLAAVLC--RQRRE
MUL_0949|M.ulcerans_Agy99 ALYVRLEINET----PVFARAMAQADDQESTAAARAPLAAVLC--RQRRE
MAV_4627|M.avium_104 GIVVRLRVEET----PAFR------ELQDLQAASTVPIRDILRERRSRRN
MLBr_00556|M.leprae_Br4923 AFAVWERCARTRLIDPAGVHIRPFLSALGASVAAGAALMVTLVNVELFGQ
: . :
Mb3503c|M.bovis_AF2122/97 LLLCFLVTLGGTVAFYTYSVNAPAIVKSVYGSQAMTATWINLVGLILLMM
Rv3476c|M.tuberculosis_H37Rv LLLCFLVTLGGTVAFYTYSVNAPAIVKSVYGSQAMTATWINLVGLILLMM
Mflv_2381|M.gilvum_PYR-GCK ILQIIGLVVIHNVGFYIVYTFLPSYFTETLDFSKVDAFWSITVASLVGIV
Mvan_4266|M.vanbaalenii_PYR-1 ILQIIGLVVVHNVGFYVVYTFLPSYFTRTLEFSKIDAFYSITVASLVGIV
MSMEG_4889|M.smegmatis_MC2_155 ILQVIGMFIVFNVGYYVVFTFIPTYFIKTLEFSKTESFVSITLASLTALV
TH_3722|M.thermoresistible__bu ILQVIGLMIIFNVAYYVVFAYMPTYLIKELGLSSSDAFISSTIACVVALV
MAB_2263c|M.abscessus_ATCC_199 MVVMMLLVIALNITNYTLLSYQPTYLKTRLGMSETQGQIVILIGEVAMMA
MMAR_4238|M.marinum_M IVLAAGAMLAAFGCLYLASTFLPSYAQLHLGYSRNLVLLIGVLGGLVCIG
MUL_0949|M.ulcerans_Agy99 IVLAAGAMLAAFGCLYLASTFLPSYAQLHLGYSRNLVLLIGVLGGLVCIG
MAV_4627|M.avium_104 TVLGLLSRWAEGSAFNTWGVFAISYATGALGLNRVSVLIAVTIAALLMAV
MLBr_00556|M.leprae_Br4923 GVLGMDQTQAAGLLVWFLIALPIGAVLGGWGATKAGDRTMTFVGLLITAG
: :. :
Mb3503c|M.bovis_AF2122/97 -LQPIGGMISDKIGRKPLLLWFGVGGLIYTYVLVTYLPETRSPTMSFLLV
Rv3476c|M.tuberculosis_H37Rv -LQPIGGMISDKIGRKPLLLWFGVGGLIYTYVLVTYLPETRSPTMSFLLV
Mflv_2381|M.gilvum_PYR-GCK -LIPPLGALSDRIGRKSLLIAGAVAFAVFAYPLFLLLNTGSLAAAIAAHA
Mvan_4266|M.vanbaalenii_PYR-1 -LIPPLGALSDRVGRKPLLLAGSIGFAVFAYPLFLLLNTGSLAAAIAAHA
MSMEG_4889|M.smegmatis_MC2_155 -LILPLAALSDRVGRKPLLLAGSIGFVVLAYPLFLLMTSGSVAAAIVGHC
TH_3722|M.thermoresistible__bu -LILPLAALSDRIGRKPLLVGGAIAFAVLAYPLFLLLNSGSTAAAMIAHA
MAB_2263c|M.abscessus_ATCC_199 -FLPFCGALSDRVGRKPMWAFSLTGLLVLSLPMFWLMGQG-FGWAIVGFA
MMAR_4238|M.marinum_M -FVALSAILCDRFGRRRVMLVGWAVGLFWSPLVMPLIGSGDTVGFTVALL
MUL_0949|M.ulcerans_Agy99 -FVALSAILCDRFGRRRVMLVGWAVGLFWSPLVMPLIGSGDTVGFTVALL
MAV_4627|M.avium_104 -LLPVSGVLTDRFGARRVYLAGIACYGLAAFPAFALFGTKKLLWFGLAMV
MLBr_00556|M.leprae_Br4923 GYWLISHWPVDLLNYRRSIFGLFSVPTMYADLLVAGLGLGLVIGPLSSAT
* .. : . : . :
Mb3503c|M.bovis_AF2122/97 AVGYVILTGYCSINALVKSELFPAHVRALGVGVG--YALANSVFGGTAPL
Rv3476c|M.tuberculosis_H37Rv AVGYVILTGYCSINALVKSELFPAHVRALGVGVG--YALANSVFGGTAPL
Mflv_2381|M.gilvum_PYR-GCK ALA-AIESVFVSASLAAGAELFATKVRSSGYSIG--YNVSVAVFGGTAPY
Mvan_4266|M.vanbaalenii_PYR-1 ALA-AIEAVFVCASLAAGAELFATRVRSSGYSIG--YNVSVAIFGGTAPY
MSMEG_4889|M.smegmatis_MC2_155 VLA-AIESTYVATAVSAGVELFATRVRYSGFSIG--YNISVAVFGGTTPY
TH_3722|M.thermoresistible__bu GLA-AIESVVISVAVVTAVEQFTTRVRYSGMSVG--YNVCVALFGGTTPY
MAB_2263c|M.abscessus_ATCC_199 VLG-LLYIPQLSTITATFPCMFPTQVRYAGFAIS--YNLSTAAFGGTAPL
MMAR_4238|M.marinum_M GLY-AVAASGFGPAASLIPELFATRYRYTGTALA--LNVAGIVGGAAPPL
MUL_0949|M.ulcerans_Agy99 GLY-AVAASGFGPAASLIPELFATRYRYTGTALA--LNVAGIVGGAAPPL
MAV_4627|M.avium_104 IVFGVVHALFYGAQGTLYSALYPTNTRYTGLSFV--YQFSGVYASGVTPM
MLBr_00556|M.leprae_Br4923 LRVVPTAQHGIASAAVVVARMTGMLIGVAALTAWGLYRFNQILAGLSTAV
. . . ..
Mb3503c|M.bovis_AF2122/97 IYQALKERDQVPMFIAYVTACIAVSLIVYVFFIKNKADTYLDREQGFAFY
Rv3476c|M.tuberculosis_H37Rv IYQALKERDQVPMFIAYVTACIAVSLIVYVFFIKNKADTYLDREQGFAFY
Mflv_2381|M.gilvum_PYR-GCK VATWLVSRTGNDIAPAYYLIAAAIVSLLTVLTMRETARKPLRKLVHP---
Mvan_4266|M.vanbaalenii_PYR-1 VATWLVSRTGNDIAPAFYVIAAAVVTFLTVLTMRETARQPLRKLVSD---
MSMEG_4889|M.smegmatis_MC2_155 VVTWLTAATGNHHAPALYLIVAAVVSVAAVLTLTENAGRPLPTGHADDAQ
TH_3722|M.thermoresistible__bu VMTWLIDTTANPLAPACAVTLAAVVSLGVVMTLRETAARPLAG-------
MAB_2263c|M.abscessus_ATCC_199 VNDLVVARTGWDLFPAAYLMVACVVGLCALPFLIETAGASIAGTEIPCNE
MMAR_4238|M.marinum_M IAGTLLASYGSSAVGLMMTALVAASLLCTYLLPETAGAPLTAA-------
MUL_0949|M.ulcerans_Agy99 IAGTLLASYGSSAVGLMMTALVAASLLCTYLLPETAGAPLTAA-------
MAV_4627|M.avium_104 ILTALIAALGG-APWLACGYLVATAVISVLATALIRDRDLYL--------
MLBr_00556|M.leprae_Br4923 PPDATLIERAAAVADQAKRAYTMMYSDIFMITAIVCVIGALLGLLISSRK
Mb3503c|M.bovis_AF2122/97 GHA-------
Rv3476c|M.tuberculosis_H37Rv GHA-------
Mflv_2381|M.gilvum_PYR-GCK ----------
Mvan_4266|M.vanbaalenii_PYR-1 ----------
MSMEG_4889|M.smegmatis_MC2_155 RATMTS----
TH_3722|M.thermoresistible__bu ----TP----
MAB_2263c|M.abscessus_ATCC_199 AAVPTDTVRA
MMAR_4238|M.marinum_M ----------
MUL_0949|M.ulcerans_Agy99 ----------
MAV_4627|M.avium_104 ----------
MLBr_00556|M.leprae_Br4923 EHASEPKVPE