For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTAQPEPAVWQGHTRGSSDYRRLLAALFCAGVATFAQLYSPQAVLPLIASDLGTGAAHAALAISAATIGL AFGVLPWAALADRVGRVQVITVSVVVATVLGLLVPFAPTYALLLCGRFVEGLALAGVPAVAVAYLTEEIN AGHAARAAGTYVAGTTIGGLAGRLVTGPVAEFAGWRVGVLTVALLCGMAALAFVKLAPAAQGFTPARTHW DLGRRLLTNLRSPRQLALFAQGFLLMGGFVAVYNFLGFRLSAAPFNLPQTVVSLVFLAYLAGTWASARAG AEATRFGRKPVLLVSIATMIAGVAVTMSTNAVVVLVGLVIATAGFFGAHAIASGWVGAEAGDSKAQASAL YNLFYYGGSSAVGWLGGLAFDAAGWSAVAGTVMGLAALAGLIAFALAR*
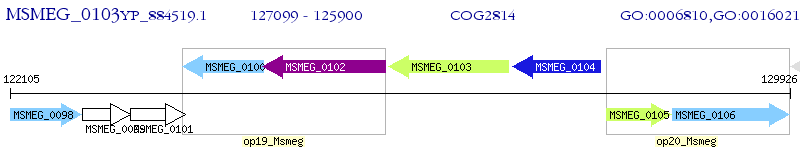
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0103 | - | - | 100% (399) | major facilitator family protein transporter |
| M. smegmatis MC2 155 | MSMEG_2810 | - | 9e-20 | 26.02% (392) | major facilitator superfamily protein |
| M. smegmatis MC2 155 | MSMEG_2028 | - | 1e-18 | 25.94% (374) | chloramphenicol resistance protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1699c | - | 2e-13 | 26.70% (412) | integral membrane transport protein |
| M. gilvum PYR-GCK | Mflv_1840 | - | 3e-16 | 25.12% (406) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv1672c | - | 2e-13 | 26.70% (412) | integral membrane transport protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2706c | - | 1e-98 | 48.69% (382) | putative transporter |
| M. marinum M | MMAR_3524 | - | 3e-14 | 23.59% (390) | hypothetical protein MMAR_3524 |
| M. avium 104 | MAV_1539 | - | 4e-87 | 45.43% (383) | major facilitator family protein transporter |
| M. thermoresistible (build 8) | TH_3714 | - | 4e-11 | 25.50% (353) | PUTATIVE major facilitator superfamily MFS_1 |
| M. ulcerans Agy99 | MUL_1468 | - | 2e-12 | 26.92% (182) | integral membrane transport protein |
| M. vanbaalenii PYR-1 | Mvan_4890 | - | 2e-18 | 26.57% (414) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1840|M.gilvum_PYR-GCK ------MSAAVDQQSTPGASVAAPDPVVRRLAVLALALGGFGIGTTEFVA
Mvan_4890|M.vanbaalenii_PYR-1 ------MTDTLHRPSGP-VSVAAPDPVVRRLAVVALALGGFGIGTTEFVA
Mb1699c|M.bovis_AF2122/97 ------------MATIAASPTHNALGKAARRLLPLLFVLYVINFVDRANI
Rv1672c|M.tuberculosis_H37Rv ------------MATIAASPTHNALGKAARRLLPLLFVLYVINFVDRANI
MAB_2706c|M.abscessus_ATCC_199 MATAASTEGTTTVRNGVDPSPHQTGTAGYRRLTGALFAAGLATFAALYGT
MAV_1539|M.avium_104 ----------MVMPNG--PVAHQAGTRGYRRMTAALCGAGLASFAAMYCT
MSMEG_0103|M.smegmatis_MC2_155 --------MVTAQPEPAVWQGHTRGSSDYRRLLAALFCAGVATFAQLYSP
MUL_1468|M.ulcerans_Agy99 ------------------------MNRTQLLTIFATGLGLFMIFLDAFIV
MMAR_3524|M.marinum_M ----------------------MKASAPSRRIVAVLLCVVPLNQIPMDAY
TH_3714|M.thermoresistible__bu ------MSAEAATPTSRVTPRRVAVASFIGTTVEFYDFLIYGTAAALVFP
Mflv_1840|M.gilvum_PYR-GCK MGLLPDIARSFDVSEPVAGHVISAYALGVVVGAPLIAAITARMARRTLLL
Mvan_4890|M.vanbaalenii_PYR-1 MGLLPDIATGMGVSEPTAGHVISAYALGVVVGAPVIAAVTARMARRKLLL
Mb1699c|M.bovis_AF2122/97 SVAALAMNADLRLSATAYGTAAGVFFLGYVLFQVPANAALARFGAGRTLT
Rv1672c|M.tuberculosis_H37Rv SVAALAMNADLRLSATAYGTAAGVFFLGYVLFQVPANAALARFGAGRTLT
MAB_2706c|M.abscessus_ATCC_199 QAVLPALSADLGVSPASAALTVSVTTGMLALSIIPASALSERFGRTRVML
MAV_1539|M.avium_104 QALLPALSAHHRIGPATAALTVSLTTGALALSIIPASVLSERYGRTRVML
MSMEG_0103|M.smegmatis_MC2_155 QAVLPLIASDLGTGAAHAALAISAATIGLAFGVLPWAALADRVGRVQVIT
MUL_1468|M.ulcerans_Agy99 NVALPDIQNTFGVGEDGLQWVVAAYSLGMAVFIMSAATLADRYGRRRWYL
MMAR_3524|M.marinum_M TPALPQMETSIGTTATALQSTVTVFMIGMALSYLAVGIMSDSWGRKPVLM
TH_3714|M.thermoresistible__bu KLFFPTASPASGVLLSFATFGVGFFAR--PLGGIVFGHFGDRLGRKRMLV
. .
Mflv_1840|M.gilvum_PYR-GCK ALMAVFTLGNLASMLASSYETL--------VGARFLAGLPHGAYFGVAAL
Mvan_4890|M.vanbaalenii_PYR-1 ALMALFTIGNLASMLAPTYETL--------IAARFLAGLPHGAYFGVAAL
Mb1699c|M.bovis_AF2122/97 AVVLAWGVCSAATALVTSAHTL--------YLARFALGVAEGGFFPGVIA
Rv1672c|M.tuberculosis_H37Rv AVVLAWGVCSAATALVTSAHTL--------YLARFALGVAEGGFFPGVIA
MAB_2706c|M.abscessus_ATCC_199 VSAIATTIIGLLLPASPTFAVL--------LIGRAAEGAALAGIPAVAMA
MAV_1539|M.avium_104 ISGVASSVIGLLLPFSPSLGVL--------VFGRAVQGVALAGIPAVAMA
MSMEG_0103|M.smegmatis_MC2_155 VSVVVATVLGLLVPFAPTYALL--------LCGRFVEGLALAGVPAVAVA
MUL_1468|M.ulcerans_Agy99 MGIVLFTLGSIACGLAPSIAML--------TAARGVQGFGAATVSVTSLA
MMAR_3524|M.marinum_M GCAAALTVTSLACAAANNVAML--------LVLRFLQGGACSAFIVVAIA
TH_3714|M.thermoresistible__bu YSLVGMGVATVLIGLLPTYAQIGMAAPALLTLLRLAQGFAMGGEWGGATL
: . : * * .
Mflv_1840|M.gilvum_PYR-GCK VAAHLMG-PQHRAKAVAHVLTGLTVATVLGVPVASWLG----------QS
Mvan_4890|M.vanbaalenii_PYR-1 VAAHLMG-PQNRAKAVAHVLTGLTVATVLGVPIASWLG----------QS
Mb1699c|M.bovis_AF2122/97 YLTVWFP-CAQRARAVATFLLAIPVANTVGLPLS----GLIVGHVHMAGL
Rv1672c|M.tuberculosis_H37Rv YLTVWFP-CAQRARAVATFLLAIPVANTVGLPLS----GLIVGHVHMAGL
MAB_2706c|M.abscessus_ATCC_199 FLAEEVH-PGSLGSAMGRYIAGTTVGGLAGRLVA----SSVL------DV
MAV_1539|M.avium_104 LLAEEVD-ASSLGSAMGRYIAGTTIGGLAGRIVP----SVVV------QV
MSMEG_0103|M.smegmatis_MC2_155 YLTEEIN-AGHAARAAGTYVAGTTIGGLAGRLVT----GPVA------EF
MUL_1468|M.ulcerans_Agy99 LVSAAFPNPKEKARAIGIWTAVASVGTATGPTLGG----------LLVDT
MMAR_3524|M.marinum_M IAADCFE-GARLRSVNGLLGAAWSAAPILAPAVGG----------FIVEY
TH_3714|M.thermoresistible__bu MAVEHAS--ADRKGFYGAFPQMGAPAGTATATLAFFAVSRLPD--GQFLS
. . . :
Mflv_1840|M.gilvum_PYR-GCK LGWRAAFGLVVAVGVLTLTALALWLPVQLRLMRSTSP-----LTELGALR
Mvan_4890|M.vanbaalenii_PYR-1 LGWRAAFGLVVGVGLVTLTALWCWLPFQLKFMRATSP-----LTELGALR
Mb1699c|M.bovis_AF2122/97 PGWRAMFVIEALPALLLAPLLRRLLPDNPQRASWLTPEERAELSARLTED
Rv1672c|M.tuberculosis_H37Rv PGWRAMFVIEALPALLLAPLLRRLLPDNPQRASWLTPEERAELSARLTED
MAB_2706c|M.abscessus_ATCC_199 SNWRIALLGSGAMTLVCTVLFAALLPKSQFFVPKKVRVGTTLRILGVHLR
MAV_1539|M.avium_104 GTWRVALLACSLITLAGTAVFAVLVPRSRFFTPKPASVRAALRNLAGHLR
MSMEG_0103|M.smegmatis_MC2_155 AGWRVGVLTVALLCGMAALAFVKLAPAAQGFTPARTHWDLGRRLLT-NLR
MUL_1468|M.ulcerans_Agy99 WGWRSIFYVNVPMGVFVILLTVGFVAES---------------------R
MMAR_3524|M.marinum_M ASWRYVFVLIAALSVVVGLAVTWALPETLDQESRTRFELGQTWQVLTATA
TH_3714|M.thermoresistible__bu WGWRLPFLLSAVLIGIGLAVRLTLTESPDFAAVQARSAVHRMPIAEAFRR
** .
Mflv_1840|M.gilvum_PYR-GCK RPQVWLALAVGMIGFGGMFAVYTYIATTMTDVAG-MPRALVPVAL-----
Mvan_4890|M.vanbaalenii_PYR-1 RPQVWLALLVGMIGFGGMFAVYTYITTTMTDVAG-MPRGLVPLAL-----
Mb1699c|M.bovis_AF2122/97 TPAPTGRSSGAGWDLVLFAVVYGGLYFALYALQFFLPQLVASLAHGTATL
Rv1672c|M.tuberculosis_H37Rv TPAPTGRSSGAGWDLVLFAVVYGGLYFALYALQFFLPQLVASLAHGTATL
MAB_2706c|M.abscessus_ATCC_199 NPVLLIMFGLAFLLMGGFVTVYNYLGYRLVAPPFGLPTAIAGLLF-----
MAV_1539|M.avium_104 NPVLAKLFAVGFVLMGGFVTVYNYLGYRLAARPFGLAPSVVGLLF-----
MSMEG_0103|M.smegmatis_MC2_155 SPRQLALFAQGFLLMGGFVAVYNFLGFRLSAAPFNLPQTVVSLVF-----
MUL_1468|M.ulcerans_Agy99 NARAGQLDLSGQALFIVTVGAFVYAVIEGPQIGWTSPLILTLLMT-----
MMAR_3524|M.marinum_M RNRVFLALAAIFGLLAGPQLSFGVAAPFLYQDEMGFSPSAYGLIALVVG-
TH_3714|M.thermoresistible__bu HRRQILLVAGAYLSQGVFAYICVAYLVSYGTSVAGIDRTATLFGVS----
.
Mflv_1840|M.gilvum_PYR-GCK -------MVFGLGMFFGNLVGGRLADRSVVRALYL-SMGALCAMLTLFVA
Mvan_4890|M.vanbaalenii_PYR-1 -------MMFGLGMVLGNLVGGRLADGSVVRALYL-SLGALCGALALFVV
Mb1699c|M.bovis_AF2122/97 TAATLAALPYGVAALAMLAWSHRSIDRSGAQAGHI-TLPTTAAGSAALGA
Rv1672c|M.tuberculosis_H37Rv TAATLAALPYGVAALAMLAWSHRSIDRSGAQAGHI-TLPTTAAGSAALGA
MAB_2706c|M.abscessus_ATCC_199 -------LLYLAGTMSSAVAGRLADRRGRGSVLIA-AIVITAAGLALT--
MAV_1539|M.avium_104 -------LLYLVGTGTSVVAGRLADRRGRPLVLGA-ALPIAVAGLLLT--
MSMEG_0103|M.smegmatis_MC2_155 -------LAYLAGTWASARAGAEATRFGRKPVLLV-SIATMIAGVAVT--
MUL_1468|M.ulcerans_Agy99 ---------AIVGCALFLWLEGRSADPMMDLTLFR-DSSYALS-------
MMAR_3524|M.marinum_M -------VAVLLGSFSTGMLATRMAFRRLAFADWT-LYMVGAALLLGSAP
TH_3714|M.thermoresistible__bu -------VAAVLAVVMYPLFGSLSDTFGRKPLFLAGVIAMGVSIGPAFAL
. .
Mflv_1840|M.gilvum_PYR-GCK ASHNPWTALLALFGIGLTGS---AVGPALQTRLMDVAHDAQTLAAALNHS
Mvan_4890|M.vanbaalenii_PYR-1 ASHNPWTALLVLFLIGLTGS---AVGPALQTRLMDVAHDAQTLAAALNHS
Mb1699c|M.bovis_AF2122/97 A-LSPMSPIVTLSWLTIAVAGILAAMPAFWSRCTAALAGPRVAVAIATVN
Rv1672c|M.tuberculosis_H37Rv A-LSPMSPIVTLSWLTIAVAGILAAMPAFWSRCTAALAGPRVAVAIATVN
MAB_2706c|M.abscessus_ATCC_199 ---IPNSLLTVIVGVGVFTAGFFAAHTVASGWVGAVAQRDRAEASALYLF
MAV_1539|M.avium_104 ---VPPSLAAIVAGVGVFTGGFFAAHTVASGWVGAVAQRDRAEASALYLF
MSMEG_0103|M.smegmatis_MC2_155 ---MSTNAVVVLVGLVIATAGFFGAHAIASGWVGAEAGDSKAQASALYNL
MUL_1468|M.ulcerans_Agy99 --------IGTICTVFFAVYGMLLLTTQFLHNVRGYTQGSGVVD------
MMAR_3524|M.marinum_M VVGVNPWAITLAVSLAVAGCG-ALVPQAQAAALGAFSRNLGLVSGLFGTL
TH_3714|M.thermoresistible__bu IDTGRPALFLVALVLVFGLAMAPAAGVTGALFAMTFDADVRYSGVSIGYT
: .
Mflv_1840|M.gilvum_PYR-GCK ALNIGNATGA--WVGGLVIAAGYGYTAPAGAGAVLALAGLVVLTVSVVLQ
Mvan_4890|M.vanbaalenii_PYR-1 ALNIGNATGA--WVGGLVIAAGLGYTAPAAAGAVLALAGLAVLTVSVLLQ
Mb1699c|M.bovis_AF2122/97 AVASLASFAGPYATGHLKDATGTYHLALLTVAAVLAAAAACSLLLRHAGR
Rv1672c|M.tuberculosis_H37Rv AVASLASFAGPYATGHLKDATGTYHLALLTVAAVLAAAAACSLLLRHAGR
MAB_2706c|M.abscessus_ATCC_199 SYYLGSSVAG--AVGGLAYQAGGWGLTVLFVGALLAIALVLAALMVLRN-
MAV_1539|M.avium_104 SYYLGGSVAG--AFGGVLYGVGGWSATVCFVVVLLMAGAALVALLVRDNG
MSMEG_0103|M.smegmatis_MC2_155 FYYGGSSAVG--WLGGLAFDAAGWSAVAGTVMGLAALAGLIAFALAR---
MUL_1468|M.ulcerans_Agy99 --------------------------------------------------
MMAR_3524|M.marinum_M TYLIIAAIMAIVGVSPERTQAPLGWLYVLCGALVFAALAWVTPRVQGEKP
TH_3714|M.thermoresistible__bu LSQVLGSAFAPTIAAALYAATESSGSIVAYLIGVSVISAVSVCLLPGGWG
Mflv_1840|M.gilvum_PYR-GCK KR--------------------
Mvan_4890|M.vanbaalenii_PYR-1 KRG-------------------
Mb1699c|M.bovis_AF2122/97 TVCANDSEIMLHPSPATPFV--
Rv1672c|M.tuberculosis_H37Rv TVCANDSEIMLHPSPATPFV--
MAB_2706c|M.abscessus_ATCC_199 -RFGHINAPE------------
MAV_1539|M.avium_104 FRIGRRVVAGVASVK-------
MSMEG_0103|M.smegmatis_MC2_155 ----------------------
MUL_1468|M.ulcerans_Agy99 ----------------------
MMAR_3524|M.marinum_M TPG-------------------
TH_3714|M.thermoresistible__bu RTGDQPATEVPASVRPMVSGPD