For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTDTLHRPSGPVSVAAPDPVVRRLAVVALALGGFGIGTTEFVAMGLLPDIATGMGVSEPTAGHVISAYAL GVVVGAPVIAAVTARMARRKLLLALMALFTIGNLASMLAPTYETLIAARFLAGLPHGAYFGVAALVAAHL MGPQNRAKAVAHVLTGLTVATVLGVPIASWLGQSLGWRAAFGLVVGVGLVTLTALWCWLPFQLKFMRATS PLTELGALRRPQVWLALLVGMIGFGGMFAVYTYITTTMTDVAGMPRGLVPLALMMFGLGMVLGNLVGGRL ADGSVVRALYLSLGALCGALALFVVASHNPWTALLVLFLIGLTGSAVGPALQTRLMDVAHDAQTLAAALN HSALNIGNATGAWVGGLVIAAGLGYTAPAAAGAVLALAGLAVLTVSVLLQKRG
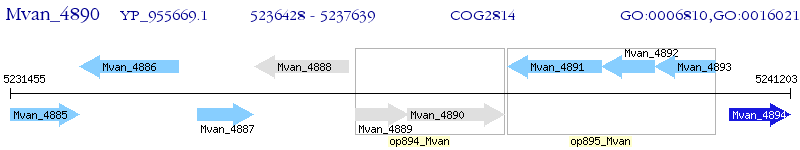
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4890 | - | - | 100% (403) | major facilitator superfamily transporter |
| M. vanbaalenii PYR-1 | Mvan_3349 | - | 5e-15 | 25.83% (391) | major facilitator superfamily transporter |
| M. vanbaalenii PYR-1 | Mvan_4475 | - | 8e-13 | 25.94% (397) | Bcr/CflA subfamily drug resistance transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0197 | - | 4e-32 | 29.82% (399) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_1840 | - | 0.0 | 88.46% (390) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv0191 | - | 4e-32 | 29.82% (399) | integral membrane protein |
| M. leprae Br4923 | MLBr_00556 | - | 8e-09 | 26.54% (162) | putative integral membrane drug transport protein |
| M. abscessus ATCC 19977 | MAB_1429 | - | 1e-144 | 65.08% (398) | major facilitator superfamily permease |
| M. marinum M | MMAR_0434 | - | 2e-30 | 28.39% (384) | integral membrane protein |
| M. avium 104 | MAV_4987 | - | 1e-29 | 27.37% (380) | sugar transporter family protein |
| M. smegmatis MC2 155 | MSMEG_5556 | - | 0.0 | 81.06% (396) | MFS permease |
| M. thermoresistible (build 8) | TH_3988 | - | 1e-30 | 33.33% (303) | PUTATIVE putative MFS permease |
| M. ulcerans Agy99 | MUL_1084 | - | 2e-30 | 28.39% (384) | integral membrane protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_4890|M.vanbaalenii_PYR-1 ---MTDTLHRPSGP-VSVAAPDPVVRRLAVVALALGGFGIGTTEFVAMGL
Mflv_1840|M.gilvum_PYR-GCK ---MSAAVDQQSTPGASVAAPDPVVRRLAVLALALGGFGIGTTEFVAMGL
MSMEG_5556|M.smegmatis_MC2_155 ---MTTSLQ-PNRP-ASVVATDTTVRWLAVFALALGGFGIGTAEFVAMGL
MAB_1429|M.abscessus_ATCC_1997 ---MTAVVDSRVQP---AAPVSPRVRRGAILALALGGFGIGTTEFVAMGL
Mb0197|M.bovis_AF2122/97 -------MTAPTGTSATTTRPWTPRIATQLSVLACAAFIYVTAEILPVGA
Rv0191|M.tuberculosis_H37Rv -------MTAPTGTSATTTRPWTPRIATQLSVLACAAFIYVTAEILPVGA
MMAR_0434|M.marinum_M -------MTSEARTATTAARPWTPRVAAQLSVLAAAAFIYVTAEILPVGA
MUL_1084|M.ulcerans_Agy99 -------MTSEARTATTAARPWTPRVAAQLSVLAAAAFIYVTAEILPVGA
MAV_4987|M.avium_104 -------MTAET--ANAAARTWTPRIAAQLAILAAAAFTYVTAEILPVGA
TH_3988|M.thermoresistible__bu MSESKASQDASAPAADPVDATARRTPWIAILAAALGIFVMITIEELPIGV
MLBr_00556|M.leprae_Br4923 -------------------MSTRAGRRVAISAGSLAVLLGALDTYVVVTI
: : . : : :
Mvan_4890|M.vanbaalenii_PYR-1 LPDIATGMGVSEPTAG---HVISAYALGVVVGAPVIAAVTARMARRKLLL
Mflv_1840|M.gilvum_PYR-GCK LPDIARSFDVSEPVAG---HVISAYALGVVVGAPLIAAITARMARRTLLL
MSMEG_5556|M.smegmatis_MC2_155 LPDIATGFGITEPTAG---HVISAYALGVVVGAPVLAAVTARVPRRVLLL
MAB_1429|M.abscessus_ATCC_1997 LPNIAHGLHVTEPQAG---HVISAYALGVVVGAPVIAALTARASRRTLLV
Mb0197|M.bovis_AF2122/97 LSAIARNLRVSVVLVG---TLLSWYALVAAVTTVPLVRWTAHWPRRRALV
Rv0191|M.tuberculosis_H37Rv LSAIARNLRVSVVLVG---TLLSWYALVAAVTTVPLVRWTAHWPRRRALV
MMAR_0434|M.marinum_M LSAIARNLHISVVLVG---TLLSWYALVAALTTVPLVRWTAHWPRRHALM
MUL_1084|M.ulcerans_Agy99 LSAIARNLHISVVLVG---TLLSWYALVAALTTVPLVRWTAHWPRRHALM
MAV_4987|M.avium_104 LPAIARNLQVSLVLVG---TLLSWYALVAALTTIPLVRWTAHLPRRRVLV
TH_3988|M.thermoresistible__bu LTLIGDDLGLSSGVVG---LTVTIPGVLAGAVAVLTPTLIGRLDRRLALV
MLBr_00556|M.leprae_Br4923 MRDIMHDVGIPVNQMQRITWIVTMYLLGYIAAMPLLSRASDRFGRKLLLQ
: * .. :. :: : : *: *
Mvan_4890|M.vanbaalenii_PYR-1 ALMALFTIGNLASMLA---PTYETLIAARFLAGLPHGAYFGVAALVAAHL
Mflv_1840|M.gilvum_PYR-GCK ALMAVFTLGNLASMLA---SSYETLVGARFLAGLPHGAYFGVAALVAAHL
MSMEG_5556|M.smegmatis_MC2_155 GLMAVFTLGNVASMVA---PSYPTLVVARFVAGLPHGAYFGVAALAAAHL
MAB_1429|M.abscessus_ATCC_1997 ALMVAFTIGNAATVFA---PGYAALIASRFVAGLPHGAYFGVAALVAAHL
Mb0197|M.bovis_AF2122/97 VSLVCLTVSQLVSALA---PNFAVLAAGRVLCAVTHGLLWAVIAPIATRL
Rv0191|M.tuberculosis_H37Rv VSLVCLTVSQLVSALA---PNFAVLAAGRVLCAVTHGLLWAVIAPIATRL
MMAR_0434|M.marinum_M VSLVCLTISQLISALA---PNFAVLAAGRALCAITHGLLWSVIAPIATRL
MUL_1084|M.ulcerans_Agy99 VSLVCLTISQLISALA---PNFAVLAAGRALCAITHGLLWSVIAPIATRL
MAV_4987|M.avium_104 ASLTCLTASQLISALA---PNFAVLAAGRVLCAITHGLLWSVIAPIATRL
TH_3988|M.thermoresistible__bu SALLLVFVSAATSAIA---PNLPVLLASRLFAGMAIGVFWALLAVVVARV
MLBr_00556|M.leprae_Br4923 VSLAGFAIGSVMTALAGQFGDFHMLIAGRTIQGVASGALLPITLALGADL
: . . : .* * .* . .:. * : : :
Mvan_4890|M.vanbaalenii_PYR-1 MGPQNRAKAVAHVLTGLTVATVLGVPIASWLGQSLG-WRAAFGLVVGVGL
Mflv_1840|M.gilvum_PYR-GCK MGPQHRAKAVAHVLTGLTVATVLGVPVASWLGQSLG-WRAAFGLVVAVGV
MSMEG_5556|M.smegmatis_MC2_155 MGPANRAKAVAHVLSGLTIATVLGVPMASWLGQALG-WRSAFGLVVFVGV
MAB_1429|M.abscessus_ATCC_1997 AGPGERAKAVGQTMLGLSVANVVGVPVATWLGNTVG-WRSAFAVVAVIGA
Mb0197|M.bovis_AF2122/97 VPPSHAGRATTSIYIGTSLALVVGSPLTAAMSLMWG-WRLAAVCVTGAAA
Rv0191|M.tuberculosis_H37Rv VPPSHAGRATTSIYIGTSLALVVGSPLTAAMSLMWG-WRLAAVCVTGAAA
MMAR_0434|M.marinum_M VPPSHAGRATTSIYIGTSLALVIGSPLTAALSLMWG-WRLAAVCVTVAAA
MUL_1084|M.ulcerans_Agy99 VPPSHAGRATTSIYIGTSLALVIGSPLTAALSLMWG-WRLAAVCVTVAAA
MAV_4987|M.avium_104 VPPSHAGRATMSIYVGTSLALVVGSPLTAALSLMWG-WRLAVVCVTVAAA
TH_3988|M.thermoresistible__bu AHPDDLPKALTLAFGGVSVAVVLGVPFAAWIGAMVG-WRWAFVLVGALGL
MLBr_00556|M.leprae_Br4923 WAQRNRAGVLGGIGAAQELGSVLGPLYGIFIVWLFSDWRYVFWINIPLTA
. . . :. *:* : . ** .
Mvan_4890|M.vanbaalenii_PYR-1 VTLTALWCWLPFQLKFMRATSPLTELG-ALRRPQ----------------
Mflv_1840|M.gilvum_PYR-GCK LTLTALALWLPVQLRLMRSTSPLTELG-ALRRPQ----------------
MSMEG_5556|M.smegmatis_MC2_155 LTLAALWFWLPEQLRTMHISSPLTELG-ALRRPQ----------------
MAB_1429|M.abscessus_ATCC_1997 ITIAALLRYLPG-LTDMRITNPLTELG-ALRRVQ----------------
Mb0197|M.bovis_AF2122/97 AVALAARLALPEMVLRADQLEHVGRRARHHRNPR----------------
Rv0191|M.tuberculosis_H37Rv AVALAARLALPEMVLRADQLEHVGRRARHHRNPR----------------
MMAR_0434|M.marinum_M VVTVAARLLLPEMVLSADQLQYVGPRSRHHRNRA----------------
MUL_1084|M.ulcerans_Agy99 VVTVAARLLLPEMVLSADQLQHVGPRSRHHRNRA----------------
MAV_4987|M.avium_104 VVTVAARLMLPEMVLTEHQLAHVGPRSRHHRNGR----------------
TH_3988|M.thermoresistible__bu MVAVALTVLLPPVPVAERTT--LGQLGAAWSIPA----------------
MLBr_00556|M.leprae_Br4923 IAMLMIQVSLSAHDRGDELEKVDVVGGVLLAIALGLVVIGLYNPQPDSKQ
. *. .
Mvan_4890|M.vanbaalenii_PYR-1 -----------------------------------------VWLALLVGM
Mflv_1840|M.gilvum_PYR-GCK -----------------------------------------VWLALAVGM
MSMEG_5556|M.smegmatis_MC2_155 -----------------------------------------VWLAVLVGM
MAB_1429|M.abscessus_ATCC_1997 -----------------------------------------VWFTLLIGI
Mb0197|M.bovis_AF2122/97 -----------------------------------------LVKVSVLTM
Rv0191|M.tuberculosis_H37Rv -----------------------------------------LVKVSVLTM
MMAR_0434|M.marinum_M -----------------------------------------LIIVSLITM
MUL_1084|M.ulcerans_Agy99 -----------------------------------------LIIVSLITM
MAV_4987|M.avium_104 -----------------------------------------LITVSLLAM
TH_3988|M.thermoresistible__bu -----------------------------------------ARTGVLVTL
MLBr_00556|M.leprae_Br4923 VLPSYGVPVLVGGIVATVAFAVWERCARTRLIDPAGVHIRPFLSALGASV
:
Mvan_4890|M.vanbaalenii_PYR-1 IGFGGMFAVYTYITTTMTDVAGMPRGLVPLALMMFGLGMVLGNLVGGRLA
Mflv_1840|M.gilvum_PYR-GCK IGFGGMFAVYTYIATTMTDVAGMPRALVPVALMVFGLGMFFGNLVGGRLA
MSMEG_5556|M.smegmatis_MC2_155 IGFGGMFAVYTYISTTMTDVAGLSRGLVPLALMVFGFGMVIGNLLGGRLA
MAB_1429|M.abscessus_ATCC_1997 VGFGGMFAFYTYISTTLTSVSGLPRALVPLALSLYGIGMVTGNVLGGWMA
Mb0197|M.bovis_AF2122/97 IAVTGHFVSYTYIVVIIRDVVGVRGPNLAWLLAAYGVAGLVSVPLVARPL
Rv0191|M.tuberculosis_H37Rv IAVTGHFVSYTYIVVIIRDVVGVRGPNLAWLLAAYGVAGLVSVPLVARPL
MMAR_0434|M.marinum_M VGVTGHFVSYTYIVVIIRQVVGVRGPSLAWLLAAYGVAGVVAVALVARPL
MUL_1084|M.ulcerans_Agy99 VGVTGHFVSYTYIVVIIRQVVGVRGPSLAWLLAAYGVAGVVAVALVARPL
MAV_4987|M.avium_104 VAVTGHFVSYTFIVEIIRNVVGVRGPTLAWVLAAYGLAGLLSVPLVARPL
TH_3988|M.thermoresistible__bu VMVTAHFTAYTYASPVLQDRIHITESGVGPMLFVFGIAGFLGNFAAGPLL
MLBr_00556|M.leprae_Br4923 AAGAALMVTLVNVELFGQGVLGMDQTQAAGLLVWFLIALPIGAVLGGWGA
. :. . : * : .. . .
Mvan_4890|M.vanbaalenii_PYR-1 DGSVVRALYLSLGALCGALALFV---VASH----NPWTALLV----LFLI
Mflv_1840|M.gilvum_PYR-GCK DRSVVRALYLSMGALCAMLTLFV---AASH----NPWTALLA----LFGI
MSMEG_5556|M.smegmatis_MC2_155 DLSVVRALYVSLAALGILLAVFV---AASH----HPWSALLV----LFGI
MAB_1429|M.abscessus_ATCC_1997 DRSVVRAIAVGLVLVAAMLLIFA---AVSR----NPWAALAL----VFAV
Mb0197|M.bovis_AF2122/97 DRWPKGAVIVGMTGLTAAFTLLT---ALAFGERHTAATALLG-TGAIVLW
Rv0191|M.tuberculosis_H37Rv DRWPKGAVIVGMTGLTAAFTLLT---ALAFGERHTAATALLG-TGAIVLW
MMAR_0434|M.marinum_M DRRPKGTIIFCVAGLTFAFVLLT---ALAFGGHLAPMTALVVGTGAIVLW
MUL_1084|M.ulcerans_Agy99 DRRPKGTIIFCVAGLTFAFVLLT---ALAFGGHLAPMTALVVGTGAIVLW
MAV_4987|M.avium_104 DHRPKSAVILCMTGLTAAFAVLT---ALAFGGPTGATTALIG-TAAIVLW
TH_3988|M.thermoresistible__bu RRSMRATYVVLPIGVMVGVXXXG---PNSR-------------------W
MLBr_00556|M.leprae_Br4923 TKAGDRTMTFVGLLITAGGYWLISHWPVDLLNYRRSIFGLFSVPTMYADL
: . :
Mvan_4890|M.vanbaalenii_PYR-1 GLTGSAVGPALQTRLMDVAHDAQTLAAALNHSALNIGNATGAWVGGLVIA
Mflv_1840|M.gilvum_PYR-GCK GLTGSAVGPALQTRLMDVAHDAQTLAAALNHSALNIGNATGAWVGGLVIA
MSMEG_5556|M.smegmatis_MC2_155 GVAGSAVGPALQTRLMDVAHDAQTLAAALNHSALNIGNATGAWVGGLVIA
MAB_1429|M.abscessus_ATCC_1997 GVSGTSLVPALQTRLMDVADDAQTLAAALNHSALNIANAGGAWLGGLVIT
Mb0197|M.bovis_AF2122/97 GALATAVSPMLQSAAMRSGGDDPDGASGLYVTAFQIGIMAGALLGGLLYE
Rv0191|M.tuberculosis_H37Rv GALATAVSPMLQSAAMRSGGDDPDGASGLYVTAFQIGIMAGALLGGLLYE
MMAR_0434|M.marinum_M GAAATAVSPMLQSAAMRSGADDPDGASGLYVTAFQVGIMAGSLIGGLLYE
MUL_1084|M.ulcerans_Agy99 GAAVTAVSPMLQSAAMRSGADDPDGASGLYVTAFQVGIMAGSLIGGLLYE
MAV_4987|M.avium_104 GAMATAVSPMMQSAAMRNGADDPDGASGLYVTAFQVGIMAGSLLGGLLYE
TH_3988|M.thermoresistible__bu RPDGTRM-------------------------------------------
MLBr_00556|M.leprae_Br4923 LVAGLGLGLVIGPLSSATLRVVPTAQHGIASAAVVVARMTGMLIGVAALT
:
Mvan_4890|M.vanbaalenii_PYR-1 AG----------------------------------LGYTAPAAAG-AVL
Mflv_1840|M.gilvum_PYR-GCK AG----------------------------------YGYTAPAGAG-AVL
MSMEG_5556|M.smegmatis_MC2_155 AG----------------------------------FGYTAPAAAG-AVL
MAB_1429|M.abscessus_ATCC_1997 AG----------------------------------YGYRAPSLVG-AAL
Mb0197|M.bovis_AF2122/97 RS----------------------------------LAMMLTASAGLMGV
Rv0191|M.tuberculosis_H37Rv RS----------------------------------LAMMLTASAGLMGV
MMAR_0434|M.marinum_M RS----------------------------------VAMMLTASAGLMGV
MUL_1084|M.ulcerans_Agy99 RS----------------------------------VAMMLTASAGLMGV
MAV_4987|M.avium_104 RS----------------------------------VILMLSASGVLMAV
TH_3988|M.thermoresistible__bu --------------------------------------------------
MLBr_00556|M.leprae_Br4923 AWGLYRFNQILAGLSTAVPPDATLIERAAAVADQAKRAYTMMYSDIFMIT
Mvan_4890|M.vanbaalenii_PYR-1 ALAGLAVLTVSVLLQKRG----------
Mflv_1840|M.gilvum_PYR-GCK ALAGLVVLTVSVVLQKR-----------
MSMEG_5556|M.smegmatis_MC2_155 AVGGLLVLTLSVAVQRRSKT--------
MAB_1429|M.abscessus_ATCC_1997 AIAGLVVLGLSVLYARSRAVALPSRAG-
Mb0197|M.bovis_AF2122/97 ALFGMTVSQHLFENPTLSPGDG------
Rv0191|M.tuberculosis_H37Rv ALFGMTVSQHLFENPTLSPGDG------
MMAR_0434|M.marinum_M ALVGMTSARRVFEAPVAVAGATSHNQ--
MUL_1084|M.ulcerans_Agy99 ALVGMTSARRVFEAPVAVAGATSHNQ--
MAV_4987|M.avium_104 ALVGIAANRRMLDVAPTSSRDS------
TH_3988|M.thermoresistible__bu ----------------------------
MLBr_00556|M.leprae_Br4923 AIVCVIGALLGLLISSRKEHASEPKVPE