For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ATAASTEGTTTVRNGVDPSPHQTGTAGYRRLTGALFAAGLATFAALYGTQAVLPALSADLGVSPASAALT VSVTTGMLALSIIPASALSERFGRTRVMLVSAIATTIIGLLLPASPTFAVLLIGRAAEGAALAGIPAVAM AFLAEEVHPGSLGSAMGRYIAGTTVGGLAGRLVASSVLDVSNWRIALLGSGAMTLVCTVLFAALLPKSQF FVPKKVRVGTTLRILGVHLRNPVLLIMFGLAFLLMGGFVTVYNYLGYRLVAPPFGLPTAIAGLLFLLYLA GTMSSAVAGRLADRRGRGSVLIAAIVITAAGLALTIPNSLLTVIVGVGVFTAGFFAAHTVASGWVGAVAQ RDRAEASALYLFSYYLGSSVAGAVGGLAYQAGGWGLTVLFVGALLAIALVLAALMVLRNRFGHINAPE*
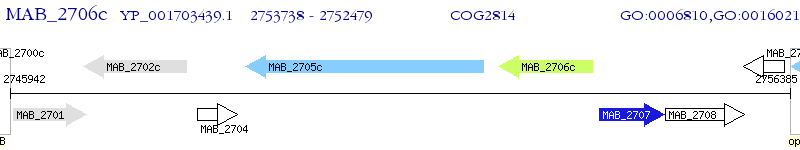
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2706c | - | - | 100% (419) | putative transporter |
| M. abscessus ATCC 19977 | MAB_2038 | - | 4e-57 | 31.65% (376) | putative transport protein |
| M. abscessus ATCC 19977 | MAB_0022c | - | 7e-23 | 26.17% (386) | permease |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0197 | - | 1e-17 | 24.72% (356) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_2728 | - | 1e-14 | 26.65% (394) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv0191 | - | 1e-17 | 24.72% (356) | integral membrane protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_0434 | - | 1e-20 | 25.31% (399) | integral membrane protein |
| M. avium 104 | MAV_1539 | - | 1e-145 | 67.51% (394) | major facilitator family protein transporter |
| M. smegmatis MC2 155 | MSMEG_0103 | - | 1e-98 | 48.69% (382) | major facilitator family protein transporter |
| M. thermoresistible (build 8) | TH_3988 | - | 1e-14 | 26.21% (290) | PUTATIVE putative MFS permease |
| M. ulcerans Agy99 | MUL_1084 | - | 6e-21 | 25.31% (399) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_1083 | - | 1e-16 | 28.06% (310) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_2706c|M.abscessus_ATCC_199 MATAASTEGTTTVRNGVDPSPHQTGTAGYRRLTGALFAAG--LATFAALY
MAV_1539|M.avium_104 ----------MVMPNG--PVAHQAGTRGYRRMTAALCGAG--LASFAAMY
MSMEG_0103|M.smegmatis_MC2_155 --------MVTAQPEPAVWQGHTRGSSDYRRLLAALFCAG--VATFAQLY
Mb0197|M.bovis_AF2122/97 ----------MTAPTGTSATTTRPWTPRIATQLSVLACAA--FIYVTAEI
Rv0191|M.tuberculosis_H37Rv ----------MTAPTGTSATTTRPWTPRIATQLSVLACAA--FIYVTAEI
MMAR_0434|M.marinum_M ----------MTSEARTATTAARPWTPRVAAQLSVLAAAA--FIYVTAEI
MUL_1084|M.ulcerans_Agy99 ----------MTSEARTATTAARPWTPRVAAQLSVLAAAA--FIYVTAEI
TH_3988|M.thermoresistible__bu -----MSESKASQDASAPAADPVDATARRTPWIAILAAALGIFVMITIEE
Mflv_2728|M.gilvum_PYR-GCK ------------------------MLRSQPKAVWAVAFAS--VVAFMGIG
Mvan_1083|M.vanbaalenii_PYR-1 ------------------MNTTPPPLSRTAAAMTVVAMGLGYTAAIVDPT
: . .
MAB_2706c|M.abscessus_ATCC_199 GTQAVLPALSADLGVSPASAALTVSVTTGMLALSIIPASALSERFGRTRV
MAV_1539|M.avium_104 CTQALLPALSAHHRIGPATAALTVSLTTGALALSIIPASVLSERYGRTRV
MSMEG_0103|M.smegmatis_MC2_155 SPQAVLPLIASDLGTGAAHAALAISAATIGLAFGVLPWAALADRVGRVQV
Mb0197|M.bovis_AF2122/97 LPVGALSAIARNLRVSVVLVGTLLSWYALVAAVTTVPLVRWTAHWPRRRA
Rv0191|M.tuberculosis_H37Rv LPVGALSAIARNLRVSVVLVGTLLSWYALVAAVTTVPLVRWTAHWPRRRA
MMAR_0434|M.marinum_M LPVGALSAIARNLHISVVLVGTLLSWYALVAALTTVPLVRWTAHWPRRHA
MUL_1084|M.ulcerans_Agy99 LPVGALSAIARNLHISVVLVGTLLSWYALVAALTTVPLVRWTAHWPRRHA
TH_3988|M.thermoresistible__bu LPIGVLTLIGDDLGLSSGVVGLTVTIPGVLAGAVAVLTPTLIGRLDRRLA
Mflv_2728|M.gilvum_PYR-GCK LVDPILKPIADNLDASPSQVSLLFTSYMAVMGVAMLITGVVSSRIGPKRT
Mvan_1083|M.vanbaalenii_PYR-1 ILSAEMSTVRIGLGMSTSAASFTASLATLTMAAAVLGAGALGDVYGMRRM
: : . .. : . :
MAB_2706c|M.abscessus_ATCC_199 MLVSAIATTIIGLLLPASPTFAVLLIGRAAEGAALAGIPAVAMAFLAEEV
MAV_1539|M.avium_104 MLISGVASSVIGLLLPFSPSLGVLVFGRAVQGVALAGIPAVAMALLAEEV
MSMEG_0103|M.smegmatis_MC2_155 ITVSVVVATVLGLLVPFAPTYALLLCGRFVEGLALAGVPAVAVAYLTEEI
Mb0197|M.bovis_AF2122/97 LVVSLVCLTVSQLVSALAPNFAVLAAGRVLCAVTHGLLWAVIAPIATRLV
Rv0191|M.tuberculosis_H37Rv LVVSLVCLTVSQLVSALAPNFAVLAAGRVLCAVTHGLLWAVIAPIATRLV
MMAR_0434|M.marinum_M LMVSLVCLTISQLISALAPNFAVLAAGRALCAITHGLLWSVIAPIATRLV
MUL_1084|M.ulcerans_Agy99 LMVSLVCLTISQLISALAPNFAVLAAGRALCAITHGLLWSVIAPIATRLV
TH_3988|M.thermoresistible__bu LVSALLLVFVSAATSAIAPNLPVLLASRLFAGMAIGVFWALLAVVVARVA
Mflv_2728|M.gilvum_PYR-GCK LLLGLVIIIAGAGLAGTSDSVMGIVGWRALWGLGNALFIATALATIVSSA
Mvan_1083|M.vanbaalenii_PYR-1 YVAGLLGTIAFGVLGAAAPTAAVLMVARAGLGVALAFLMGLSLAIVNAVS
. : : . : * . . . .
MAB_2706c|M.abscessus_ATCC_199 HPGSLGSAMGRYIAGTTVGGLAGRLVASSVLDVSNWRIALLGSGAMTLVC
MAV_1539|M.avium_104 DASSLGSAMGRYIAGTTIGGLAGRIVPSVVVQVGTWRVALLACSLITLAG
MSMEG_0103|M.smegmatis_MC2_155 NAGHAARAAGTYVAGTTIGGLAGRLVTGPVAEFAGWRVGVLTVALLCGMA
Mb0197|M.bovis_AF2122/97 PPSHAGRATTSIYIGTSLALVVGSPLTAAMSLMWGWRLAAVCVTGAAAAV
Rv0191|M.tuberculosis_H37Rv PPSHAGRATTSIYIGTSLALVVGSPLTAAMSLMWGWRLAAVCVTGAAAAV
MMAR_0434|M.marinum_M PPSHAGRATTSIYIGTSLALVIGSPLTAALSLMWGWRLAAVCVTVAAAVV
MUL_1084|M.ulcerans_Agy99 PPSHAGRATTSIYIGTSLALVIGSPLTAALSLMWGWRLAAVCVTVAAAVV
TH_3988|M.thermoresistible__bu HPDDLPKALTLAFGGVSVAVVLGVPFAAWIGAMVGWRWAFVLVGALGLMV
Mflv_2728|M.gilvum_PYR-GCK KG-SVAQAIILYEAALGVGIAVGPLVGGVLGSVS-WRGPFFGVSVLMVVA
Mvan_1083|M.vanbaalenii_PYR-1 APERRTTVIAWYFGVGFAVAAPLPAIGGPLATQMGWRAALLVTPALAAVA
. . . : ** .
MAB_2706c|M.abscessus_ATCC_199 TVLFAALLPKSQFFVPKKVRVGTTLRIL----------------------
MAV_1539|M.avium_104 TAVFAVLVPRSRFFTPKPASVRAALRNL----------------------
MSMEG_0103|M.smegmatis_MC2_155 ALAFVKLAPAAQGFTPARTHWDLGRRLL----------------------
Mb0197|M.bovis_AF2122/97 ALAARLALPEMVLRADQLEHVGRRA-------------------------
Rv0191|M.tuberculosis_H37Rv ALAARLALPEMVLRADQLEHVGRRA-------------------------
MMAR_0434|M.marinum_M TVAARLLLPEMVLSADQLQYVGPRS-------------------------
MUL_1084|M.ulcerans_Agy99 TVAARLLLPEMVLSADQLQHVGPRS-------------------------
TH_3988|M.thermoresistible__bu AVALTVLLPP--VPVAERTTLGQLG-------------------------
Mflv_2728|M.gilvum_PYR-GCK LVVTAFLLPATPPAARATTLADPFR-------------------------
Mvan_1083|M.vanbaalenii_PYR-1 LALTLRYVPHIPRTHRALDLPGLALAAVALLALVFGIARLETAGDVVAVT
*
MAB_2706c|M.abscessus_ATCC_199 -------------------------GVHLRNPVLLIMFGLAFLLMGGFVT
MAV_1539|M.avium_104 -------------------------AGHLRNPVLAKLFAVGFVLMGGFVT
MSMEG_0103|M.smegmatis_MC2_155 -------------------------T-NLRSPRQLALFAQGFLLMGGFVA
Mb0197|M.bovis_AF2122/97 --------------------------RHHRNPRLVKVSVLTMIAVTGHFV
Rv0191|M.tuberculosis_H37Rv --------------------------RHHRNPRLVKVSVLTMIAVTGHFV
MMAR_0434|M.marinum_M --------------------------RHHRNRALIIVSLITMVGVTGHFV
MUL_1084|M.ulcerans_Agy99 --------------------------RHHRNRALIIVSLITMVGVTGHFV
TH_3988|M.thermoresistible__bu --------------------------AAWSIPAARTGVLVTLVMVTAHFT
Mflv_2728|M.gilvum_PYR-GCK ---------------------------ALRHRGLFGVGVTALLYNFGFFT
Mvan_1083|M.vanbaalenii_PYR-1 ALGTAVLAAGGFVAWELRTAQPALDLSVFRSGRFNAAVTAGVVFNFLTGG
.:
MAB_2706c|M.abscessus_ATCC_199 VYNYLGYRLVAPPFGLPTAIAGLLFLLYLAGTMSSAVAGRLADRRGRGSV
MAV_1539|M.avium_104 VYNYLGYRLAARPFGLAPSVVGLLFLLYLVGTGTSVVAGRLADRRGRPLV
MSMEG_0103|M.smegmatis_MC2_155 VYNFLGFRLSAAPFNLPQTVVSLVFLAYLAGTWASARAGAEATRFGRKPV
Mb0197|M.bovis_AF2122/97 SYTYIVVIIRDVVGVRGPNLAWLLAAYGVAGLVSVPLVARPLDRWPKGAV
Rv0191|M.tuberculosis_H37Rv SYTYIVVIIRDVVGVRGPNLAWLLAAYGVAGLVSVPLVARPLDRWPKGAV
MMAR_0434|M.marinum_M SYTYIVVIIRQVVGVRGPSLAWLLAAYGVAGVVAVALVARPLDRRPKGTI
MUL_1084|M.ulcerans_Agy99 SYTYIVVIIRQVVGVRGPSLAWLLAAYGVAGVVAVALVARPLDRRPKGTI
TH_3988|M.thermoresistible__bu AYTYASPVLQDRIHITESGVGPMLFVFGIAGFLGNFAAGPLLRRSMRATY
Mflv_2728|M.gilvum_PYR-GCK LLAFTPFPLDMGAQQIGLIFFGWGVALAVTSVFVAPRLQRRFGTVPTLLV
Mvan_1083|M.vanbaalenii_PYR-1 LVLLFAFYLVTVRGESPEVLGLLLIPATVLAAVAATTAGPAAVRLGDRPV
: . : .
MAB_2706c|M.abscessus_ATCC_199 LIAAIVITAAGLALTIPNS---------LLTVIVGVGVFTAGFFAAHTVA
MAV_1539|M.avium_104 LGAALPIAVAGLLLTVPPS---------LAAIVAGVGVFTGGFFAAHTVA
MSMEG_0103|M.smegmatis_MC2_155 LLVSIATMIAGVAVTMSTN---------AVVVLVGLVIATAGFFGAHAIA
Mb0197|M.bovis_AF2122/97 IVGMTGLTAAFTLLTALAFGERHTAATALLG-TGAIVLWGALATAVSPML
Rv0191|M.tuberculosis_H37Rv IVGMTGLTAAFTLLTALAFGERHTAATALLG-TGAIVLWGALATAVSPML
MMAR_0434|M.marinum_M IFCVAGLTFAFVLLTALAFGGHLAPMTALVVGTGAIVLWGAAATAVSPML
MUL_1084|M.ulcerans_Agy99 IFCVAGLTFAFVLLTALAFGGHLAPMTALVVGTGAIVLWGAAVTAVSPML
TH_3988|M.thermoresistible__bu VVLPIGVMVGVXXX------------------------------------
Mflv_2728|M.gilvum_PYR-GCK NLIAMSATLAVMAVGADHK----------AVLAACVVVAGLWIGINNTLI
Mvan_1083|M.vanbaalenii_PYR-1 LVGGLAVMLAAVLLLRLFDENTSLTVVFTAVALMVVGGALVATPQASIMM
.
MAB_2706c|M.abscessus_ATCC_199 SGWVGAVAQRDRAEASALYLFSYYLGSSVAGAVGGLAYQAG---------
MAV_1539|M.avium_104 SGWVGAVAQRDRAEASALYLFSYYLGGSVAGAFGGVLYGVG---------
MSMEG_0103|M.smegmatis_MC2_155 SGWVGAEAGDSKAQASALYNLFYYGGSSAVGWLGGLAFDAA---------
Mb0197|M.bovis_AF2122/97 QSAAMRSGGDDPDGASGLYVTAFQIGIMAGALLGGLLYER----------
Rv0191|M.tuberculosis_H37Rv QSAAMRSGGDDPDGASGLYVTAFQIGIMAGALLGGLLYER----------
MMAR_0434|M.marinum_M QSAAMRSGADDPDGASGLYVTAFQVGIMAGSLIGGLLYER----------
MUL_1084|M.ulcerans_Agy99 QSAAMRSGADDPDGASGLYVTAFQVGIMAGSLIGGLLYER----------
TH_3988|M.thermoresistible__bu -------GPNSRWRPDGTRM------------------------------
Mflv_2728|M.gilvum_PYR-GCK TEAVMKAAPVERGVASAAYSFMRFGGAAVAPWLAGVLGETF---------
Mvan_1083|M.vanbaalenii_PYR-1 AAAPAHLGGAVSGVKSAVNQTGYSLGPTVFALVGINLILAEGTEKLAGSG
. ..
MAB_2706c|M.abscessus_ATCC_199 ----GWGLTVLFVGALLAIALVLAALMVLRN--RFGHINAPE--------
MAV_1539|M.avium_104 ----GWSATVCFVVVLLMAGAALVALLVRDNGFRIGRRVVAGVASVK---
MSMEG_0103|M.smegmatis_MC2_155 ----GWSAVAGTVMGLAALAGLIAFALAR---------------------
Mb0197|M.bovis_AF2122/97 ------SLAMMLTASAGLMGVALFGMTVSQHLFENPTLSPGDG-------
Rv0191|M.tuberculosis_H37Rv ------SLAMMLTASAGLMGVALFGMTVSQHLFENPTLSPGDG-------
MMAR_0434|M.marinum_M ------SVAMMLTASAGLMGVALVGMTSARRVFEAPVAVAGATSHNQ---
MUL_1084|M.ulcerans_Agy99 ------SVAMMLTASAGLMGVALVGMTSARRVFEAPVAVAGATSHNQ---
TH_3988|M.thermoresistible__bu --------------------------------------------------
Mflv_2728|M.gilvum_PYR-GCK ----SVHVPFWVGAGAVLLAAGALAATRTHLRHLDDEETGIDELTDEARA
Mvan_1083|M.vanbaalenii_PYR-1 ITLEEAREAFRATHGGSAGGSHLLDPDRAQIVATAATESMLDAIHTISLL
MAB_2706c|M.abscessus_ATCC_199 ------------------------
MAV_1539|M.avium_104 ------------------------
MSMEG_0103|M.smegmatis_MC2_155 ------------------------
Mb0197|M.bovis_AF2122/97 ------------------------
Rv0191|M.tuberculosis_H37Rv ------------------------
MMAR_0434|M.marinum_M ------------------------
MUL_1084|M.ulcerans_Agy99 ------------------------
TH_3988|M.thermoresistible__bu ------------------------
Mflv_2728|M.gilvum_PYR-GCK LTVGSDS-----------------
Mvan_1083|M.vanbaalenii_PYR-1 MAAIPVVAIVVALIWLKPAPRHQV