For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PVAIWVLGAAIFAQGTSELMLAGLLPEVSADFAVSIPQAGLTVSVFALGMLVGAPILAIGTLRWPRRRAL LAFLGVFVVAHIVAALTDSYAVLLATRFVAAFVYAGFWAVGASTAMALVSPARRGRAMSVVAGGITVAMV VGLPAGTWIGQHFGWRAAVWTVTVLTLVAAGAVLTAVPDTRAHTVMRARDEIRGVAVPRLWLSYGLTATS TAALLGTFTYLSAMLITTTGVRADWVPLVLLGYGLGSLAGMAGGGFAADRWPRGVLAAGFGMLTVVLALL SVTASVPAAVASLAVVLGFAGFGTNPALNSRVFGIAPGAPTLASAGNISAFNVGISAGPWLAGLALTAGL GYPSVPWIGAGLGVVALLLLAVDVLVGRAPRRAAEPVGVAVGCG*
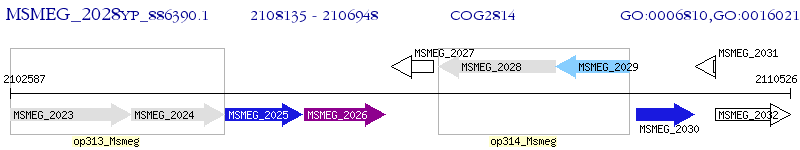
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2028 | - | - | 100% (395) | chloramphenicol resistance protein |
| M. smegmatis MC2 155 | MSMEG_6245 | - | 4e-67 | 39.53% (387) | chloramphenicol resistance protein |
| M. smegmatis MC2 155 | MSMEG_5556 | - | 7e-60 | 33.33% (378) | MFS permease |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0197 | - | 2e-36 | 31.38% (376) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_1840 | - | 2e-58 | 34.39% (378) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv0191 | - | 2e-36 | 31.38% (376) | integral membrane protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2983c | - | 6e-66 | 38.06% (381) | chloramphenicol resistance protein |
| M. marinum M | MMAR_0577 | - | 2e-33 | 30.25% (367) | integral membrane transport protein |
| M. avium 104 | MAV_4987 | - | 6e-32 | 30.31% (353) | sugar transporter family protein |
| M. thermoresistible (build 8) | TH_3988 | - | 1e-19 | 28.12% (256) | PUTATIVE putative MFS permease |
| M. ulcerans Agy99 | MUL_1084 | - | 9e-32 | 28.91% (377) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_4890 | - | 2e-59 | 34.04% (376) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0197|M.bovis_AF2122/97 -------MTAPTGTSATTTRPWTPRIATQLSVLACAAFIYVTAEILPVGA
Rv0191|M.tuberculosis_H37Rv -------MTAPTGTSATTTRPWTPRIATQLSVLACAAFIYVTAEILPVGA
MUL_1084|M.ulcerans_Agy99 -------MTSEARTATTAARPWTPRVAAQLSVLAAAAFIYVTAEILPVGA
MAV_4987|M.avium_104 -------MTAET--ANAAARTWTPRIAAQLAILAAAAFTYVTAEILPVGA
MSMEG_2028|M.smegmatis_MC2_155 -------------------------MPVAIWVLGAAIFAQGTSELMLAGL
MAB_2983c|M.abscessus_ATCC_199 -------------------------MTLPIYALTLAIFAQGCSEFMFAGL
Mflv_1840|M.gilvum_PYR-GCK ---MSAAVDQQSTPGASVAAPDPVVRRLAVLALALGGFGIGTTEFVAMGL
Mvan_4890|M.vanbaalenii_PYR-1 ---MTDTLHRPSGP-VSVAAPDPVVRRLAVVALALGGFGIGTTEFVAMGL
MMAR_0577|M.marinum_M ----------MPESVTAPPELGIRHATPGLLALTIATCLAITTEILPVGL
TH_3988|M.thermoresistible__bu MSESKASQDASAPAADPVDATARRTPWIAILAAALGIFVMITIEELPIGV
: . * : *
Mb0197|M.bovis_AF2122/97 LSAIARNLRVSVVLVGTLLSWYALVAAVTTVPLVRWTAHWPRRRALVVSL
Rv0191|M.tuberculosis_H37Rv LSAIARNLRVSVVLVGTLLSWYALVAAVTTVPLVRWTAHWPRRRALVVSL
MUL_1084|M.ulcerans_Agy99 LSAIARNLHISVVLVGTLLSWYALVAALTTVPLVRWTAHWPRRHALMVSL
MAV_4987|M.avium_104 LPAIARNLQVSLVLVGTLLSWYALVAALTTIPLVRWTAHLPRRRVLVASL
MSMEG_2028|M.smegmatis_MC2_155 LPEVSADFAVSIPQAGLTVSVFALGMLVGAPILAIGTLRWPRRRALLAFL
MAB_2983c|M.abscessus_ATCC_199 ITEVGDDFGVSLAAAGTLTSAFAVGIIVGAPLMALISGRWPRRRALVLFL
Mflv_1840|M.gilvum_PYR-GCK LPDIARSFDVSEPVAGHVISAYALGVVVGAPLIAAITARMARRTLLLALM
Mvan_4890|M.vanbaalenii_PYR-1 LPDIATGMGVSEPTAGHVISAYALGVVVGAPVIAAVTARMARRKLLLALM
MMAR_0577|M.marinum_M LPAIGHTFGVRDAVTGLLVSLYAVLVALLAVPLTVVTARFPRKPLLLTTL
TH_3988|M.thermoresistible__bu LTLIGDDLGLSSGVVGLTVTIPGVLAGAVAVLTPTLIGRLDRRLALVSAL
:. :. : : .* : .: : : *: *: :
Mb0197|M.bovis_AF2122/97 VCLTVSQLVSALAPNFAVLAAGRVLCAVTHGLLWAVIAPIATRLVPPSHA
Rv0191|M.tuberculosis_H37Rv VCLTVSQLVSALAPNFAVLAAGRVLCAVTHGLLWAVIAPIATRLVPPSHA
MUL_1084|M.ulcerans_Agy99 VCLTISQLISALAPNFAVLAAGRALCAITHGLLWSVIAPIATRLVPPSHA
MAV_4987|M.avium_104 TCLTASQLISALAPNFAVLAAGRVLCAITHGLLWSVIAPIATRLVPPSHA
MSMEG_2028|M.smegmatis_MC2_155 GVFVVAHIVAALTDSYAVLLATRFVAAFVYAGFWAVGASTAMALVSPARR
MAB_2983c|M.abscessus_ATCC_199 SVFIAAHIVGATAASFPLLLIVRVVSAVANAGFLAVALTTAMSITGPRAK
Mflv_1840|M.gilvum_PYR-GCK AVFTLGNLASMLASSYETLVGARFLAGLPHGAYFGVAALVAAHLMGPQHR
Mvan_4890|M.vanbaalenii_PYR-1 ALFTIGNLASMLAPTYETLIAARFLAGLPHGAYFGVAALVAAHLMGPQNR
MMAR_0577|M.marinum_M LGYAVSNTLVAIAPTFAMVAAGRAVGGVTHALFFSLCIGYAPRLVGLADV
TH_3988|M.thermoresistible__bu LLVFVSAATSAIAPNLPVLLASRLFAGMAIGVFWALLAVVVARVAHPDDL
. : . : * . .. . .: . :
Mb0197|M.bovis_AF2122/97 GRATTSIYIGTSLALVVGSPLTAAMSLMWGWRLAAVCVTGAAAAVALAAR
Rv0191|M.tuberculosis_H37Rv GRATTSIYIGTSLALVVGSPLTAAMSLMWGWRLAAVCVTGAAAAVALAAR
MUL_1084|M.ulcerans_Agy99 GRATTSIYIGTSLALVIGSPLTAALSLMWGWRLAAVCVTVAAAVVTVAAR
MAV_4987|M.avium_104 GRATMSIYVGTSLALVVGSPLTAALSLMWGWRLAVVCVTVAAAVVTVAAR
MSMEG_2028|M.smegmatis_MC2_155 GRAMSVVAGGITVAMVVGLPAGTWIGQHFGWRAAVWTVTVLTLVAAGAVL
MAB_2983c|M.abscessus_ATCC_199 GRTTALLLSGITIACVAGVPGGAALGYFWGWRAALWAVVLVSVPALAAIM
Mflv_1840|M.gilvum_PYR-GCK AKAVAHVLTGLTVATVLGVPVASWLGQSLGWRAAFGLVVAVGVLTLTALA
Mvan_4890|M.vanbaalenii_PYR-1 AKAVAHVLTGLTVATVLGVPIASWLGQSLGWRAAFGLVVGVGLVTLTALW
MMAR_0577|M.marinum_M GRALALVGGGVSAGIVVGVPLLTSVGTAVGWRGAFAVLAVLAAVLLLVVA
TH_3988|M.thermoresistible__bu PKALTLAFGGVSVAVVLGVPFAAWIGAMVGWRWAFVLVGALGLMVAVALT
:: * : . * * * : :. *** * : .
Mb0197|M.bovis_AF2122/97 LALPEMVLRADQLEHVGRRARHHRNPRLVKVSVLTMIAVTGHFVSYTYIV
Rv0191|M.tuberculosis_H37Rv LALPEMVLRADQLEHVGRRARHHRNPRLVKVSVLTMIAVTGHFVSYTYIV
MUL_1084|M.ulcerans_Agy99 LLLPEMVLSADQLQHVGPRSRHHRNRALIIVSLITMVGVTGHFVSYTYIV
MAV_4987|M.avium_104 LMLPEMVLTEHQLAHVGPRSRHHRNGRLITVSLLAMVAVTGHFVSYTFIV
MSMEG_2028|M.smegmatis_MC2_155 TAVPDTRAHT-VMRARD-EIRGVAVPRLWLSYGLTATSTAALLGTFTYLS
MAB_2983c|M.abscessus_ATCC_199 YALPVGRPVTGLPDGIG-ELRTLRNRRLQITLAAGVLCNAATFCAFTYLA
Mflv_1840|M.gilvum_PYR-GCK LWLPVQLRLMRSTSPLT-ELGALRRPQVWLALAVGMIGFGGMFAVYTYIA
Mvan_4890|M.vanbaalenii_PYR-1 CWLPFQLKFMRATSPLT-ELGALRRPQVWLALLVGMIGFGGMFAVYTYIT
MMAR_0577|M.marinum_M KLLPPVSSESAPPPAGT----AKGRRRLAAVVSSNALTYTGQFTLYTFIS
TH_3988|M.thermoresistible__bu VLLPPVPVAERTTLGQLG--AAWSIPAARTGVLVTLVMVTAHFTAYTYAS
:* . : :*:
Mb0197|M.bovis_AF2122/97 VIIRDVVGVRGPNLAWLLAAYGVAGLVSVPLVARPLDRWPKGAVIVGMTG
Rv0191|M.tuberculosis_H37Rv VIIRDVVGVRGPNLAWLLAAYGVAGLVSVPLVARPLDRWPKGAVIVGMTG
MUL_1084|M.ulcerans_Agy99 VIIRQVVGVRGPSLAWLLAAYGVAGVVAVALVARPLDRRPKGTIIFCVAG
MAV_4987|M.avium_104 EIIRNVVGVRGPTLAWVLAAYGLAGLLSVPLVARPLDHRPKSAVILCMTG
MSMEG_2028|M.smegmatis_MC2_155 AMLITTTGVRADWVPLVLLGYGLGSLAGMAGGGFAADRWPRGVLAAGFGM
MAB_2983c|M.abscessus_ATCC_199 PVSVQVTSLSPAWTPALLALFGCGSFVGVHATGRLADSHANLVAGGGGVL
Mflv_1840|M.gilvum_PYR-GCK TTMTDVAGMPRALVPVALMVFGLGMFFGNLVGGRLADRSVVRALYLSMGA
Mvan_4890|M.vanbaalenii_PYR-1 TTMTDVAGMPRGLVPLALMMFGLGMVLGNLVGGRLADGSVVRALYLSLGA
MMAR_0577|M.marinum_M AVLLAAG-ARPAFIGPLLLMCGVCSLAGLWYVGATLDRNPRQTTVIIIVV
TH_3988|M.thermoresistible__bu PVLQDRIHITESGVGPMLFVFGIAGFLGNFAAGPLLRRSMRATYVVLPIG
* * . . . .
Mb0197|M.bovis_AF2122/97 LTAAFTLLTALAFGERHTAATALLG-TGAIVLWGALATAVSPMLQSAAMR
Rv0191|M.tuberculosis_H37Rv LTAAFTLLTALAFGERHTAATALLG-TGAIVLWGALATAVSPMLQSAAMR
MUL_1084|M.ulcerans_Agy99 LTFAFVLLTALAFGGHLAPMTALVVGTGAIVLWGAAVTAVSPMLQSAAMR
MAV_4987|M.avium_104 LTAAFAVLTALAFGGPTGATTALIG-TAAIVLWGAMATAVSPMMQSAAMR
MSMEG_2028|M.smegmatis_MC2_155 LTVVLALLSVTASVPAAVASLAVVL--------GFAGFGTNPALNSRVFG
MAB_2983c|M.abscessus_ATCC_199 LLAGWIAFAGTAAHDWLFVTLVFAL--------GALSFGVGSTLFSQGLY
Mflv_1840|M.gilvum_PYR-GCK LCAMLTLFVAASHNPWTALLALFGI--------GLTGSAVGPALQTRLMD
Mvan_4890|M.vanbaalenii_PYR-1 LCGALALFVVASHNPWTALLVLFLI--------GLTGSAVGPALQTRLMD
MMAR_0577|M.marinum_M VIGALLGLGASFPALLPVVLAAAVW--------NGAFGGIPSIYQAGAVR
TH_3988|M.thermoresistible__bu VMVGVXXX------------------------------GPNSRWRPDGTR
: . . .
Mb0197|M.bovis_AF2122/97 SGGDDPDGASGLYVTAFQIGIMAGALLGGLLYER--SLAMMLTASAGLMG
Rv0191|M.tuberculosis_H37Rv SGGDDPDGASGLYVTAFQIGIMAGALLGGLLYER--SLAMMLTASAGLMG
MUL_1084|M.ulcerans_Agy99 SGADDPDGASGLYVTAFQVGIMAGSLIGGLLYER--SVAMMLTASAGLMG
MAV_4987|M.avium_104 NGADDPDGASGLYVTAFQVGIMAGSLLGGLLYER--SVILMLSASGVLMA
MSMEG_2028|M.smegmatis_MC2_155 IAPGAPTLASAGNISAFNVGISAGPWLAGLALTAGLGYPSVPWIGAGLGV
MAB_2983c|M.abscessus_ATCC_199 AASDAPMLGSSLTTAAFNVGTVIGPSLGGASIHTPLGYRGPLWVSVMLAA
Mflv_1840|M.gilvum_PYR-GCK VAHDAQTLAAALNHSALNIGNATGAWVGGLVIAAGYGYTAPAGAGAVLAL
Mvan_4890|M.vanbaalenii_PYR-1 VAHDAQTLAAALNHSALNIGNATGAWVGGLVIAAGLGYTAPAAAGAVLAL
MMAR_0577|M.marinum_M AQAATPEMAGAWVNATANLGIAAGAAIGAGLLPT-IGLAGLPWVAAALIA
TH_3988|M.thermoresistible__bu M-------------------------------------------------
Mb0197|M.bovis_AF2122/97 VALFGMTVSQHLFENPTLSPGDG-------
Rv0191|M.tuberculosis_H37Rv VALFGMTVSQHLFENPTLSPGDG-------
MUL_1084|M.ulcerans_Agy99 VALVGMTSARRVFEAPVAVAGATSHNQ---
MAV_4987|M.avium_104 VALVGIAANRRMLDVAPTSSRDS-------
MSMEG_2028|M.smegmatis_MC2_155 VALLLLAVDVLVGRAPRRAAEPVGVAVGCG
MAB_2983c|M.abscessus_ATCC_199 LALVVTLCGLQARRRTRRAVTSARSS----
Mflv_1840|M.gilvum_PYR-GCK AGLVVLTVSVVLQKR---------------
Mvan_4890|M.vanbaalenii_PYR-1 AGLAVLTVSVLLQKRG--------------
MMAR_0577|M.marinum_M ISLAVTLATGRGPTH---------------
TH_3988|M.thermoresistible__bu ------------------------------