For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SAAVDQQSTPGASVAAPDPVVRRLAVLALALGGFGIGTTEFVAMGLLPDIARSFDVSEPVAGHVISAYAL GVVVGAPLIAAITARMARRTLLLALMAVFTLGNLASMLASSYETLVGARFLAGLPHGAYFGVAALVAAHL MGPQHRAKAVAHVLTGLTVATVLGVPVASWLGQSLGWRAAFGLVVAVGVLTLTALALWLPVQLRLMRSTS PLTELGALRRPQVWLALAVGMIGFGGMFAVYTYIATTMTDVAGMPRALVPVALMVFGLGMFFGNLVGGRL ADRSVVRALYLSMGALCAMLTLFVAASHNPWTALLALFGIGLTGSAVGPALQTRLMDVAHDAQTLAAALN HSALNIGNATGAWVGGLVIAAGYGYTAPAGAGAVLALAGLVVLTVSVVLQKR*
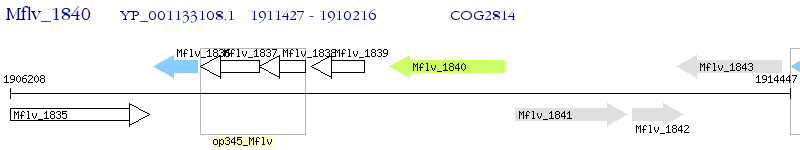
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1840 | - | - | 100% (403) | major facilitator transporter |
| M. gilvum PYR-GCK | Mflv_1730 | - | 9e-13 | 22.99% (422) | EmrB/QacA family drug resistance transporter |
| M. gilvum PYR-GCK | Mflv_2728 | - | 2e-11 | 26.22% (370) | major facilitator transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0197 | - | 1e-33 | 30.30% (396) | integral membrane protein |
| M. tuberculosis H37Rv | Rv0191 | - | 1e-33 | 30.30% (396) | integral membrane protein |
| M. leprae Br4923 | MLBr_01562 | - | 3e-05 | 25.43% (173) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | MAB_1429 | - | 1e-145 | 64.66% (399) | major facilitator superfamily permease |
| M. marinum M | MMAR_0434 | - | 2e-30 | 28.12% (384) | integral membrane protein |
| M. avium 104 | MAV_4987 | - | 7e-31 | 28.33% (360) | sugar transporter family protein |
| M. smegmatis MC2 155 | MSMEG_5556 | - | 1e-177 | 80.31% (391) | MFS permease |
| M. thermoresistible (build 8) | TH_3988 | - | 2e-30 | 31.97% (294) | PUTATIVE putative MFS permease |
| M. ulcerans Agy99 | MUL_1084 | - | 1e-30 | 28.12% (384) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_4890 | - | 0.0 | 88.46% (390) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1840|M.gilvum_PYR-GCK --------------------------------MSAAVDQQSTPGASVAAP
Mvan_4890|M.vanbaalenii_PYR-1 --------------------------------MTDTLHRPSGP-VSVAAP
MSMEG_5556|M.smegmatis_MC2_155 --------------------------------MTTSLQ-PNRP-ASVVAT
MAB_1429|M.abscessus_ATCC_1997 --------------------------------MTAVVDSRVQP---AAPV
Mb0197|M.bovis_AF2122/97 ------------------------------------MTAPTGTSATTTRP
Rv0191|M.tuberculosis_H37Rv ------------------------------------MTAPTGTSATTTRP
MMAR_0434|M.marinum_M ------------------------------------MTSEARTATTAARP
MUL_1084|M.ulcerans_Agy99 ------------------------------------MTSEARTATTAARP
MAV_4987|M.avium_104 ------------------------------------MTAET--ANAAART
TH_3988|M.thermoresistible__bu -----------------------------MSESKASQDASAPAADPVDAT
MLBr_01562|M.leprae_Br4923 MVTLYDHERAVHNWTAARSDRPAPSRLTQQVEPASERTSKYPTLLPSGRF
Mflv_1840|M.gilvum_PYR-GCK DPVVRRLAVLALALGGFGIGTTEFVAMGLLPDIARSFDVSEPVAGHVISA
Mvan_4890|M.vanbaalenii_PYR-1 DPVVRRLAVVALALGGFGIGTTEFVAMGLLPDIATGMGVSEPTAGHVISA
MSMEG_5556|M.smegmatis_MC2_155 DTTVRWLAVFALALGGFGIGTAEFVAMGLLPDIATGFGITEPTAGHVISA
MAB_1429|M.abscessus_ATCC_1997 SPRVRRGAILALALGGFGIGTTEFVAMGLLPNIAHGLHVTEPQAGHVISA
Mb0197|M.bovis_AF2122/97 WTPRIATQLSVLACAAFIYVTAEILPVGALSAIARNLRVSVVLVGTLLSW
Rv0191|M.tuberculosis_H37Rv WTPRIATQLSVLACAAFIYVTAEILPVGALSAIARNLRVSVVLVGTLLSW
MMAR_0434|M.marinum_M WTPRVAAQLSVLAAAAFIYVTAEILPVGALSAIARNLHISVVLVGTLLSW
MUL_1084|M.ulcerans_Agy99 WTPRVAAQLSVLAAAAFIYVTAEILPVGALSAIARNLHISVVLVGTLLSW
MAV_4987|M.avium_104 WTPRIAAQLAILAAAAFTYVTAEILPVGALPAIARNLQVSLVLVGTLLSW
TH_3988|M.thermoresistible__bu ARRTPWIAILAAALGIFVMITIEELPIGVLTLIGDDLGLSSGVVGLTVTI
MLBr_01562|M.leprae_Br4923 PSGRFIAAVIVIGGMQLLATMDSTVAIVALPKIQNELSLSDAGRSWGITA
: . : . :.: *. * : :: . ::
Mflv_1840|M.gilvum_PYR-GCK YALGVVVGAPLIAAITARMARRTLLLALMAVFTLGNLASMLASSYETLVG
Mvan_4890|M.vanbaalenii_PYR-1 YALGVVVGAPVIAAVTARMARRKLLLALMALFTIGNLASMLAPTYETLIA
MSMEG_5556|M.smegmatis_MC2_155 YALGVVVGAPVLAAVTARVPRRVLLLGLMAVFTLGNVASMVAPSYPTLVV
MAB_1429|M.abscessus_ATCC_1997 YALGVVVGAPVIAALTARASRRTLLVALMVAFTIGNAATVFAPGYAALIA
Mb0197|M.bovis_AF2122/97 YALVAAVTTVPLVRWTAHWPRRRALVVSLVCLTVSQLVSALAPNFAVLAA
Rv0191|M.tuberculosis_H37Rv YALVAAVTTVPLVRWTAHWPRRRALVVSLVCLTVSQLVSALAPNFAVLAA
MMAR_0434|M.marinum_M YALVAALTTVPLVRWTAHWPRRHALMVSLVCLTISQLISALAPNFAVLAA
MUL_1084|M.ulcerans_Agy99 YALVAALTTVPLVRWTAHWPRRHALMVSLVCLTISQLISALAPNFAVLAA
MAV_4987|M.avium_104 YALVAALTTIPLVRWTAHLPRRRVLVASLTCLTASQLISALAPNFAVLAA
TH_3988|M.thermoresistible__bu PGVLAGAVAVLTPTLIGRLDRRLALVSALLLVFVSAATSAIAPNLPVLLA
MLBr_01562|M.leprae_Br4923 YVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCAVAWDEVTLVI
: *: :: : . . .* .*
Mflv_1840|M.gilvum_PYR-GCK ARFLAGLPHGAYFGVAALVAAHLMGPQHRAKAVAHVLTGLTVATVLGVPV
Mvan_4890|M.vanbaalenii_PYR-1 ARFLAGLPHGAYFGVAALVAAHLMGPQNRAKAVAHVLTGLTVATVLGVPI
MSMEG_5556|M.smegmatis_MC2_155 ARFVAGLPHGAYFGVAALAAAHLMGPANRAKAVAHVLSGLTIATVLGVPM
MAB_1429|M.abscessus_ATCC_1997 SRFVAGLPHGAYFGVAALVAAHLAGPGERAKAVGQTMLGLSVANVVGVPV
Mb0197|M.bovis_AF2122/97 GRVLCAVTHGLLWAVIAPIATRLVPPSHAGRATTSIYIGTSLALVVGSPL
Rv0191|M.tuberculosis_H37Rv GRVLCAVTHGLLWAVIAPIATRLVPPSHAGRATTSIYIGTSLALVVGSPL
MMAR_0434|M.marinum_M GRALCAITHGLLWSVIAPIATRLVPPSHAGRATTSIYIGTSLALVIGSPL
MUL_1084|M.ulcerans_Agy99 GRALCAITHGLLWSVIAPIATRLVPPSHAGRATTSIYIGTSLALVIGSPL
MAV_4987|M.avium_104 GRVLCAITHGLLWSVIAPIATRLVPPSHAGRATMSIYVGTSLALVVGSPL
TH_3988|M.thermoresistible__bu SRLFAGMAIGVFWALLAVVVARVAHPDDLPKALTLAFGGVSVAVVLGVPF
MLBr_01562|M.leprae_Br4923 ARLLQGIGSAIASPTGLALVATTFSKGSARNTATAVFAAMTAIGSVMGLV
.* . .: . .: .: . : : .
Mflv_1840|M.gilvum_PYR-GCK ASWLGQSLGWRAAFGLVVAVGVLTLTALALWLPVQLRLMRSTSPLTELG-
Mvan_4890|M.vanbaalenii_PYR-1 ASWLGQSLGWRAAFGLVVGVGLVTLTALWCWLPFQLKFMRATSPLTELG-
MSMEG_5556|M.smegmatis_MC2_155 ASWLGQALGWRSAFGLVVFVGVLTLAALWFWLPEQLRTMHISSPLTELG-
MAB_1429|M.abscessus_ATCC_1997 ATWLGNTVGWRSAFAVVAVIGAITIAALLRYLPG-LTDMRITNPLTELG-
Mb0197|M.bovis_AF2122/97 TAAMSLMWGWRLAAVCVTGAAAAVALAARLALPEMVLRADQLEHVGRRAR
Rv0191|M.tuberculosis_H37Rv TAAMSLMWGWRLAAVCVTGAAAAVALAARLALPEMVLRADQLEHVGRRAR
MMAR_0434|M.marinum_M TAALSLMWGWRLAAVCVTVAAAVVTVAARLLLPEMVLSADQLQYVGPRSR
MUL_1084|M.ulcerans_Agy99 TAALSLMWGWRLAAVCVTVAAAVVTVAARLLLPEMVLSADQLQHVGPRSR
MAV_4987|M.avium_104 TAALSLMWGWRLAVVCVTVAAAVVTVAARLMLPEMVLTEHQLAHVGPRSR
TH_3988|M.thermoresistible__bu AAWIGAMVGWRWAFVLVGALGLMVAVALTVLLPPVPVAERTT--LGQLGA
MLBr_01562|M.leprae_Br4923 VGGALTEVSWRLAFLVNVPIGLVMIYLARTALHETHKERMKLDAAGAILA
. .** * . *
Mflv_1840|M.gilvum_PYR-GCK ALRRPQVWLALAVG------------------------------------
Mvan_4890|M.vanbaalenii_PYR-1 ALRRPQVWLALLVG------------------------------------
MSMEG_5556|M.smegmatis_MC2_155 ALRRPQVWLAVLVG------------------------------------
MAB_1429|M.abscessus_ATCC_1997 ALRRVQVWFTLLIG------------------------------------
Mb0197|M.bovis_AF2122/97 HHRNPRLVKVSVLT------------------------------------
Rv0191|M.tuberculosis_H37Rv HHRNPRLVKVSVLT------------------------------------
MMAR_0434|M.marinum_M HHRNRALIIVSLIT------------------------------------
MUL_1084|M.ulcerans_Agy99 HHRNRALIIVSLIT------------------------------------
MAV_4987|M.avium_104 HHRNGRLITVSLLA------------------------------------
TH_3988|M.thermoresistible__bu AWSIPAARTGVLVT------------------------------------
MLBr_01562|M.leprae_Br4923 TLACTAAVFAFSMGPEKGWISLTTIGSGLVALVAAIAFAVVERTAENPVV
:
Mflv_1840|M.gilvum_PYR-GCK -----------------MIGFGGMFAVYTYIATTMTDVAGMPRALVPVAL
Mvan_4890|M.vanbaalenii_PYR-1 -----------------MIGFGGMFAVYTYITTTMTDVAGMPRGLVPLAL
MSMEG_5556|M.smegmatis_MC2_155 -----------------MIGFGGMFAVYTYISTTMTDVAGLSRGLVPLAL
MAB_1429|M.abscessus_ATCC_1997 -----------------IVGFGGMFAFYTYISTTLTSVSGLPRALVPLAL
Mb0197|M.bovis_AF2122/97 -----------------MIAVTGHFVSYTYIVVIIRDVVGVRGPNLAWLL
Rv0191|M.tuberculosis_H37Rv -----------------MIAVTGHFVSYTYIVVIIRDVVGVRGPNLAWLL
MMAR_0434|M.marinum_M -----------------MVGVTGHFVSYTYIVVIIRQVVGVRGPSLAWLL
MUL_1084|M.ulcerans_Agy99 -----------------MVGVTGHFVSYTYIVVIIRQVVGVRGPSLAWLL
MAV_4987|M.avium_104 -----------------MVAVTGHFVSYTFIVEIIRNVVGVRGPTLAWVL
TH_3988|M.thermoresistible__bu -----------------LVMVTAHFTAYTYASPVLQDRIHITESGVGPML
MLBr_01562|M.leprae_Br4923 PFDLFRDRNRLVTFIAIFLAGGVIFSLTVSIGLYIQDILGYSALRAGIGF
:: * . : . :
Mflv_1840|M.gilvum_PYR-GCK MVFGLGMFFGNLVGGRLADRSVVRALYLS-----MGALCAMLTLFVAASH
Mvan_4890|M.vanbaalenii_PYR-1 MMFGLGMVLGNLVGGRLADGSVVRALYLS-----LGALCGALALFVVASH
MSMEG_5556|M.smegmatis_MC2_155 MVFGFGMVIGNLLGGRLADLSVVRALYVS-----LAALGILLAVFVAASH
MAB_1429|M.abscessus_ATCC_1997 SLYGIGMVTGNVLGGWMADRSVVRAIAVG-----LVLVAAMLLIFAAVSR
Mb0197|M.bovis_AF2122/97 AAYGVAGLVSVPLVARPLDRWPKGAVIVG-----MTGLTAAFTLLTALAF
Rv0191|M.tuberculosis_H37Rv AAYGVAGLVSVPLVARPLDRWPKGAVIVG-----MTGLTAAFTLLTALAF
MMAR_0434|M.marinum_M AAYGVAGVVAVALVARPLDRRPKGTIIFC-----VAGLTFAFVLLTALAF
MUL_1084|M.ulcerans_Agy99 AAYGVAGVVAVALVARPLDRRPKGTIIFC-----VAGLTFAFVLLTALAF
MAV_4987|M.avium_104 AAYGLAGLLSVPLVARPLDHRPKSAVILC-----MTGLTAAFAVLTALAF
TH_3988|M.thermoresistible__bu FVFGIAGFLGNFAAGPLLRRSMRATYVVL-----PIGVMVGVXXXGPNSR
MLBr_01562|M.leprae_Br4923 IPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLLMVAMLYGWAFMNRGVP
: .. . . . . :
Mflv_1840|M.gilvum_PYR-GCK -NPWTALLALFG----------------IGLTGSAVGPALQTRLMDVAHD
Mvan_4890|M.vanbaalenii_PYR-1 -NPWTALLVLFL----------------IGLTGSAVGPALQTRLMDVAHD
MSMEG_5556|M.smegmatis_MC2_155 -HPWSALLVLFG----------------IGVAGSAVGPALQTRLMDVAHD
MAB_1429|M.abscessus_ATCC_1997 -NPWAALALVFA----------------VGVSGTSLVPALQTRLMDVADD
Mb0197|M.bovis_AF2122/97 GERHTAATALLG-TGA---------IVLWGALATAVSPMLQSAAMRSGGD
Rv0191|M.tuberculosis_H37Rv GERHTAATALLG-TGA---------IVLWGALATAVSPMLQSAAMRSGGD
MMAR_0434|M.marinum_M GGHLAPMTALVVGTGA---------IVLWGAAATAVSPMLQSAAMRSGAD
MUL_1084|M.ulcerans_Agy99 GGHLAPMTALVVGTGA---------IVLWGAAVTAVSPMLQSAAMRSGAD
MAV_4987|M.avium_104 GGPTGATTALIG-TAA---------IVLWGAMATAVSPMMQSAAMRNGAD
TH_3988|M.thermoresistible__bu WRPDGTRM------------------------------------------
MLBr_01562|M.leprae_Br4923 YFPNLLAPIVVGGIGIGMAVVPLTLSAIAGVGFDRIGPVSAIALMLQSLG
Mflv_1840|M.gilvum_PYR-GCK AQTLAAALNHSALNIGNATGAWVG-------GLVIAAGYGYTAPAGAG-A
Mvan_4890|M.vanbaalenii_PYR-1 AQTLAAALNHSALNIGNATGAWVG-------GLVIAAGLGYTAPAAAG-A
MSMEG_5556|M.smegmatis_MC2_155 AQTLAAALNHSALNIGNATGAWVG-------GLVIAAGFGYTAPAAAG-A
MAB_1429|M.abscessus_ATCC_1997 AQTLAAALNHSALNIANAGGAWLG-------GLVITAGYGYRAPSLVG-A
Mb0197|M.bovis_AF2122/97 DPDGASGLYVTAFQIGIMAGALLG-------GLLYERSLAMMLTASAGLM
Rv0191|M.tuberculosis_H37Rv DPDGASGLYVTAFQIGIMAGALLG-------GLLYERSLAMMLTASAGLM
MMAR_0434|M.marinum_M DPDGASGLYVTAFQVGIMAGSLIG-------GLLYERSVAMMLTASAGLM
MUL_1084|M.ulcerans_Agy99 DPDGASGLYVTAFQVGIMAGSLIG-------GLLYERSVAMMLTASAGLM
MAV_4987|M.avium_104 DPDGASGLYVTAFQVGIMAGSLLG-------GLLYERSVILMLSASGVLM
TH_3988|M.thermoresistible__bu --------------------------------------------------
MLBr_01562|M.leprae_Br4923 GPLVLAVIQAVITSRTLYLGGTTGPVKFMNDAQLRALDHGYTYGLLWVAG
Mflv_1840|M.gilvum_PYR-GCK VLALAGLVVLTVSVVLQKR---------------
Mvan_4890|M.vanbaalenii_PYR-1 VLALAGLAVLTVSVLLQKRG--------------
MSMEG_5556|M.smegmatis_MC2_155 VLAVGGLLVLTLSVAVQRRSKT------------
MAB_1429|M.abscessus_ATCC_1997 ALAIAGLVVLGLSVLYARSRAVALPSRAG-----
Mb0197|M.bovis_AF2122/97 GVALFGMTVSQHLFENPTLSPGDG----------
Rv0191|M.tuberculosis_H37Rv GVALFGMTVSQHLFENPTLSPGDG----------
MMAR_0434|M.marinum_M GVALVGMTSARRVFEAPVAVAGATSHNQ------
MUL_1084|M.ulcerans_Agy99 GVALVGMTSARRVFEAPVAVAGATSHNQ------
MAV_4987|M.avium_104 AVALVGIAANRRMLDVAPTSSRDS----------
TH_3988|M.thermoresistible__bu ----------------------------------
MLBr_01562|M.leprae_Br4923 AVVIVGVAALFIGYTPEQVAYAQEVKEAVDAGEL