For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSDRVTAAVERALDDRQREATEEVERILAAAVRVMERAAPEPPRVSDIVAEAGSSNKAFYRYFAGKDDLI LAVMERGVAIVVSYLEHQMAKESSPDTKIARWIEGTLAQVADPHLISMSRAAAGQLSNWLATDREMLRPL RDLLTAPVTALGSSDVDRDVEAVYGCTVATMRRYVGSAERPGRDDIEHLVRFCLLGLGVS
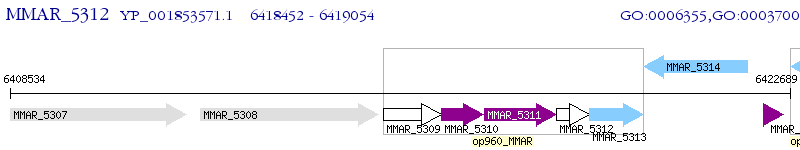
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5312 | - | - | 100% (200) | hypothetical protein MMAR_5312 |
| M. marinum M | MMAR_1569 | - | 3e-10 | 29.35% (184) | putative regulatory protein |
| M. marinum M | MMAR_0684 | - | 1e-09 | 30.07% (153) | putative regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3185c | - | 6e-05 | 23.31% (133) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_2097 | - | 8e-10 | 36.78% (87) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3160c | - | 6e-05 | 23.31% (133) | TetR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1024 | - | 1e-06 | 27.03% (148) | TetR family transcriptional regulator |
| M. avium 104 | MAV_0309 | - | 1e-92 | 84.16% (202) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_4847 | - | 2e-05 | 28.41% (88) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_3734 | - | 2e-11 | 26.24% (202) | PUTATIVE putative transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_4386 | - | 1e-106 | 97.50% (200) | hypothetical protein MUL_4386 |
| M. vanbaalenii PYR-1 | Mvan_4608 | - | 8e-09 | 33.71% (89) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3185c|M.bovis_AF2122/97 ------------------MPRQAGRWSPTALRILGAAAELIALRGYSSTS
Rv3160c|M.tuberculosis_H37Rv ------------------MPRQAGRWSPTALRILGAAAELIALRGYSSTS
MMAR_5312|M.marinum_M ------MSDRVTAAVERALDDRQREATEEVERILAAAVRVMERAAPEPPR
MUL_4386|M.ulcerans_Agy99 ------MSDRVTAAVERTLDDRQREATEEVERILAAAVRVMERAAPEPPR
MAV_0309|M.avium_104 -----MISDRVTAAVERALDDRQREATEEVERILAAAVRVMERVAPEPPR
Mflv_2097|M.gilvum_PYR-GCK ---------------------------------MEAAYRCLADPHAGPVS
Mvan_4608|M.vanbaalenii_PYR-1 -------------------MQPGPDHDGDRRRIIEAAYRCLAQPHAGPIP
TH_3734|M.thermoresistible__bu -----------------VAETRTVDRSSERQHIIDAAYRCLDRNSGSSAS
MAB_1024|M.abscessus_ATCC_1997 ---------------------MSADPDPTGERILEAALQTMLSFGIRRAT
MSMEG_4847|M.smegmatis_MC2_155 MTSAGEEPAWKQRAVERSIKTAKLRAAQRVQRFLDAAQAIIIEKGSTDFT
: ** :
Mb3185c|M.bovis_AF2122/97 TRDIAAAVGVEQPAIYKHFSAKRDILAALVRLAVEWPLELFGHITAMPV-
Rv3160c|M.tuberculosis_H37Rv TRDIAAAVGVEQPAIYKHFSAKRDILAALVRLAVEWPLELFGHITAMPV-
MMAR_5312|M.marinum_M VSDIVAEAGSSNKAFYRYFAGKDDLILAVMERGVAIVVSYLEHQMAKES-
MUL_4386|M.ulcerans_Agy99 VSDIVAEAGSSNKAFYRYFAGKDDLILAVMERGVAIVVSYLGHQMAKES-
MAV_0309|M.avium_104 VSDIVAEAGSSNKAFYRYFAGKDELILAVMERGVAIVVSYLEHQMAKEA-
Mflv_2097|M.gilvum_PYR-GCK MAAILGTAGVSSRAFYRHFESKDDLFLALLEQECSAVVGQVDRVADAAQG
Mvan_4608|M.vanbaalenii_PYR-1 MTSILRSAGLSSRAFYRHFLSKDDLFLALLRQECEAVIAQVDRIADAAVG
TH_3734|M.thermoresistible__bu ITEILDEAGLGTRAFYRNFSTKHDLLLEMFRRDREAVQAQLREAVARAP-
MAB_1024|M.abscessus_ATCC_1997 VDEIARRAGVSHMTVYRRWSNKTDLVLAVLMREAQAIFTSVDAEIAALD-
MSMEG_4847|M.smegmatis_MC2_155 VQEVVDRSRQSLRSFYLQFDGKHELLLALFEDALSRSADQIRAATASHS-
: :.* : * ::. :. .
Mb3185c|M.bovis_AF2122/97 PAVVKLHRWLTESLDHLHASPYVLVSILITPDLHQES--FVAERELVAEM
Rv3160c|M.tuberculosis_H37Rv PAVVKLHRWLTESLDHLHASPYVLVSILITPDLHQES--FVAERELVAEM
MMAR_5312|M.marinum_M SPDTKIARWIEGTLAQVADPHLISMSRAAAGQLSN---WLATDREMLRPL
MUL_4386|M.ulcerans_Agy99 RPDTKIARWIEGTLAQVADPHLISMSRAAAGQLSN---WLATQREMLRPL
MAV_0309|M.avium_104 TPRGKIARWIEGTLAQVADPHLISMTRAAAGQMSAGTSWRAADQEMMRPL
Mflv_2097|M.gilvum_PYR-GCK SPADQLAAWIEAMFDIVVDPRQRLQLTVIDSEEVRTAKGYRQTRDRLHAA
Mvan_4608|M.vanbaalenii_PYR-1 SPADQLADWIAAMFDIIVDPQQRLQLTVIDSDEVRAAKGYREMRERLHAE
TH_3734|M.thermoresistible__bu TPVEALRSWMTHMFELVSNPRKRRRVATLYSEEMRRTPGYVRELELVTAA
MAB_1024|M.abscessus_ATCC_1997 SPEEKLVAGFTGIYWHVHTHPLMRRAVETDPESVLP--VMTSGAGPALDM
MSMEG_4847|M.smegmatis_MC2_155 DPLERLKVAIQLLFESSRPDPAAKRPLFTDFAPRLLVS-HPSEVKVAHAP
. : :
Mb3185c|M.bovis_AF2122/97 ER-ALVGLIETGQGEGDVRAM-HPLSAARLVQALFDALALPEFAVSPDEI
Rv3160c|M.tuberculosis_H37Rv ER-ALVGLIETGQGEGDVRAM-HPLSAARLVQALFDALALPEFAVSPDEI
MMAR_5312|M.marinum_M RDLLTAPVTALGSSD--------VDRDVEAVY-GCTVATMRRYVGSAERP
MUL_4386|M.ulcerans_Agy99 RFLLTAPVTALGSSD--------VDRDVEAVY-GCTVATMRRYVGSAERP
MAV_0309|M.avium_104 RELLVEPVAALGSSD--------VDRDVEAVF-SCTAATLRRYVGSAQQP
Mflv_2097|M.gilvum_PYR-GCK RERGLAEILRRGLRDGSFPLT-EPEADAVAIN-AVVSRVLNTQTTDEPEA
Mvan_4608|M.vanbaalenii_PYR-1 RERSLAEILRRGRRDGSFPLA-EPEIDAVAVN-ALVSRLLVRQTTDDPQA
TH_3734|M.thermoresistible__bu DQASIAAILHRGKAEGIFTAC-TPEADARSIHAAFEAALLDRFHQKSTTD
MAB_1024|M.abscessus_ATCC_1997 ATTYLAGHVSQSAGDLVDDPYGIAEVFVRLTHSLILAPSPRRELATREDA
MSMEG_4847|M.smegmatis_MC2_155 LLALLTELMEEAGEAGLLREGINPKRMAAMTMQTVMFVAQSSGGADESTP
. .
Mb3185c|M.bovis_AF2122/97 VEFAMTALLSDPDRLAEIRAAADALEIQTAPPDRGL
Rv3160c|M.tuberculosis_H37Rv VEFAMTALLSDPDRLAEIRAAADALEIQTAPPDRGL
MMAR_5312|M.marinum_M GRDDIEHLVR--FCLLGLGVS---------------
MUL_4386|M.ulcerans_Agy99 GRDDIEHLVR--FCLLGLGVS---------------
MAV_0309|M.avium_104 APDDIAHVVR--FCLRGLGVR---------------
Mflv_2097|M.gilvum_PYR-GCK RKKALVAVLD--FALRAVGAAAR-------------
Mvan_4608|M.vanbaalenii_PYR-1 RKQAEVAVLD--FALRAVGAVPGGQDTRR-------
TH_3734|M.thermoresistible__bu AADEVEHLLA--FFLRGLGVAT--------------
MAB_1024|M.abscessus_ATCC_1997 QQYARRYILPIAQALVPAASPAVR------------
MSMEG_4847|M.smegmatis_MC2_155 NPITADEVWD--FVAHGFSADPPA------------
: