For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ISDRVTAAVERALDDRQREATEEVERILAAAVRVMERVAPEPPRVSDIVAEAGSSNKAFYRYFAGKDELI LAVMERGVAIVVSYLEHQMAKEATPRGKIARWIEGTLAQVADPHLISMTRAAAGQMSAGTSWRAADQEMM RPLRELLVEPVAALGSSDVDRDVEAVFSCTAATLRRYVGSAQQPAPDDIAHVVRFCLRGLGVR*
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
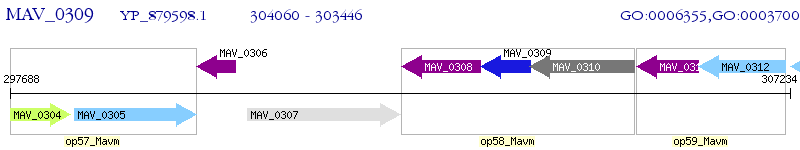
| M. avium 104 | MAV_0309 | - | - | 100% (204) | transcriptional regulator, TetR family protein |
| M. avium 104 | MAV_2439 | - | 5e-12 | 29.51% (183) | transcriptional regulator, TetR family protein, putative |
| M. avium 104 | MAV_1858 | - | 7e-08 | 31.87% (91) | transcriptional regulator, TetR family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2097 | - | 2e-09 | 24.18% (182) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3875 | - | 3e-05 | 37.88% (66) | TetR family transcriptional regulator |
| M. marinum M | MMAR_5312 | - | 2e-92 | 84.16% (202) | hypothetical protein MMAR_5312 |
| M. smegmatis MC2 155 | MSMEG_4449 | - | 7e-06 | 28.21% (117) | putative transcriptional regulator |
| M. thermoresistible (build 8) | TH_3734 | - | 8e-13 | 27.14% (199) | PUTATIVE putative transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_4386 | - | 7e-90 | 82.67% (202) | hypothetical protein MUL_4386 |
| M. vanbaalenii PYR-1 | Mvan_4608 | - | 3e-09 | 25.41% (185) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5312|M.marinum_M ---------MSDRVTAAVERALDDRQREATEEVERILAAAVRVMERAAPE
MUL_4386|M.ulcerans_Agy99 ---------MSDRVTAAVERTLDDRQREATEEVERILAAAVRVMERAAPE
MAV_0309|M.avium_104 --------MISDRVTAAVERALDDRQREATEEVERILAAAVRVMERVAPE
Mflv_2097|M.gilvum_PYR-GCK ------------------------------------MEAAYRCLADPHAG
Mvan_4608|M.vanbaalenii_PYR-1 ----------------------MQPGPDHDGDRRRIIEAAYRCLAQPHAG
TH_3734|M.thermoresistible__bu --------------------VAETRTVDRSSERQHIIDAAYRCLDRNSGS
MSMEG_4449|M.smegmatis_MC2_155 ------------MNGIEEDVMAPKRPQAAAEKRAEIVAAARRLFIDAGYD
MAB_3875|M.abscessus_ATCC_1997 MNVLPLCEHGKTMSSSARRNSEGTDRAVVRNPRVDLVSAAVRLLNEQGPD
: ** * :
MMAR_5312|M.marinum_M PPRVSDIVAEAGSSNKAFYRYFAGKDDLILAVMERGVAIVVSYLEHQMAK
MUL_4386|M.ulcerans_Agy99 PPRVSDIVAEAGSSNKAFYRYFAGKDDLILAVMERGVAIVVSYLGHQMAK
MAV_0309|M.avium_104 PPRVSDIVAEAGSSNKAFYRYFAGKDELILAVMERGVAIVVSYLEHQMAK
Mflv_2097|M.gilvum_PYR-GCK PVSMAAILGTAGVSSRAFYRHFESKDDLFLALLEQECSAVVGQVDRVADA
Mvan_4608|M.vanbaalenii_PYR-1 PIPMTSILRSAGLSSRAFYRHFLSKDDLFLALLRQECEAVIAQVDRIADA
TH_3734|M.thermoresistible__bu SASITEILDEAGLGTRAFYRNFSTKHDLLLEMFRRDREAVQAQLREAVAR
MSMEG_4449|M.smegmatis_MC2_155 ATPMSRLAKEAGVAPNTIYWYFPDKDEVLIAVL----EAVMADVWPQYEE
MAB_3875|M.abscessus_ATCC_1997 ALQTRKIAAAAGTSTMAVYTYFGGMRELIAEVAEAGLRQFAEAQAAVEQS
. : ** . :.* * ::: : .
MMAR_5312|M.marinum_M ES-SPDTKIARWIEGTLAQVADPHLISMSRAAAGQLSN---WLATDREML
MUL_4386|M.ulcerans_Agy99 ES-RPDTKIARWIEGTLAQVADPHLISMSRAAAGQLSN---WLATQREML
MAV_0309|M.avium_104 EA-TPRGKIARWIEGTLAQVADPHLISMTRAAAGQMSAGTSWRAADQEMM
Mflv_2097|M.gilvum_PYR-GCK AQGSPADQLAAWIEAMFDIVVDPRQRLQLTVIDSEEVRTAKGYRQTRDRL
Mvan_4608|M.vanbaalenii_PYR-1 AVGSPADQLADWIAAMFDIIVDPQQRLQLTVIDSDEVRAAKGYREMRERL
TH_3734|M.thermoresistible__bu AP-TPVEALRSWMTHMFELVSNPRKRRRVATLYSEEMRRTPGYVRELELV
MSMEG_4449|M.smegmatis_MC2_155 IASQPIAARLLWIVGKLTEMS-----RLVTTVHARVEHSAAVAEWHQNFH
MAB_3875|M.abscessus_ATCC_1997 DDVIADFMLTGMAYRQFAIDYPHMYRLMFGVTSAHGVNAPRRNMFDTSHV
. : . . .
MMAR_5312|M.marinum_M RPLR----------DLLTAPVTALGSSD-------VDRDVEAVY-GCTVA
MUL_4386|M.ulcerans_Agy99 RPLR----------FLLTAPVTALGSSD-------VDRDVEAVY-GCTVA
MAV_0309|M.avium_104 RPLR----------ELLVEPVAALGSSD-------VDRDVEAVF-SCTAA
Mflv_2097|M.gilvum_PYR-GCK HAAR----------ERGLAEILRRGLRDGSFPLTEPEADAVAIN-AVVSR
Mvan_4608|M.vanbaalenii_PYR-1 HAER----------ERSLAEILRRGRRDGSFPLAEPEIDAVAVN-ALVSR
TH_3734|M.thermoresistible__bu TAAD----------QASIAAILHRGKAEGIFTACTPEADARSIHAAFEAA
MSMEG_4449|M.smegmatis_MC2_155 TITG----------SLLQFELQAAGVPA-------PDAEVMIGVFTVEGL
MAB_3875|M.abscessus_ATCC_1997 TPEHPSAAYLFRCVQRAMAAGRIDGSPEDAAKVAATAAKFWTVMHGFVML
* .
MMAR_5312|M.marinum_M TMRRYVGSAERPGRDDIEHLVRFCLLGLGVS-------------------
MUL_4386|M.ulcerans_Agy99 TMRRYVGSAERPGRDDIEHLVRFCLLGLGVS-------------------
MAV_0309|M.avium_104 TLRRYVGSAQQPAPDDIAHVVRFCLRGLGVR-------------------
Mflv_2097|M.gilvum_PYR-GCK VLNTQTTDEPEARKKALVAVLDFALRAVGAAAR-----------------
Mvan_4608|M.vanbaalenii_PYR-1 LLVRQTTDDPQARKQAEVAVLDFALRAVGAVPGGQDTRR-----------
TH_3734|M.thermoresistible__bu LLDRFHQKSTTDAADEVEHLLAFFLRGLGVAT------------------
MSMEG_4449|M.smegmatis_MC2_155 LMHDHDEAQQRAIIDALASRWTTNLPRPSDLGR-----------------
MAB_3875|M.abscessus_ATCC_1997 ELAGFWGEDGAAVGPVLASMTTDLLVALGDTPERLNSSAAAAAARLAPSA
: * .