For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SCTRQHVVVTGASSGIGRSTALRLASAGWHVYAGVRRAADGEALQAASSGGQLSPLLMDVTVAEQIDAAV GIVCEHIGAAGLDGLVDNAGIGVASPVEMVPLDALRQQLEVNVVGQIAVTQAFLPLLRRAAGRIVVIASI GDRFTPPFGGPLAASKAAIATLADALRQEVAPWGIKVVMIEPASINSGAADKLERDAKKAIDEFGADGAR LYRDAYLGMVKAALSRERRGSPPTVVADLVLKVLTMARPHARNVVGKDARMLATVSRWLPTPALDALRRR VFGLPKPGSLAISRSA*
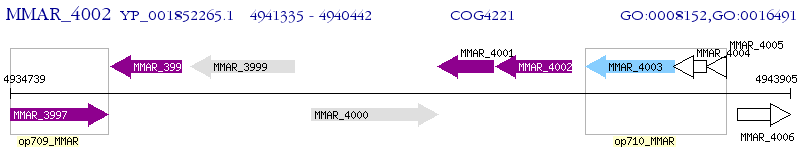
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4002 | - | - | 100% (297) | dehydrogenase/decarboxylase protein |
| M. marinum M | MMAR_3561 | - | 9e-27 | 31.03% (290) | oxidoreductase |
| M. marinum M | MMAR_1614 | - | 4e-25 | 32.16% (255) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1079 | - | 2e-18 | 35.33% (184) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_4152 | - | 2e-26 | 29.63% (297) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv1050 | - | 2e-18 | 35.33% (184) | oxidoreductase |
| M. leprae Br4923 | MLBr_00862 | ephD | 3e-16 | 31.90% (279) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1305 | - | 5e-25 | 32.50% (280) | short-chain dehydrogenase/reductase |
| M. avium 104 | MAV_2098 | - | 1e-28 | 32.50% (280) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2026 | - | 1e-25 | 33.59% (256) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_4244 | - | 6e-41 | 38.38% (284) | PUTATIVE putative Dehydrogenase/decarboxylase protein |
| M. ulcerans Agy99 | MUL_3865 | - | 1e-133 | 98.41% (251) | dehydrogenase/decarboxylase protein |
| M. vanbaalenii PYR-1 | Mvan_0982 | - | 5e-22 | 33.60% (250) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4002|M.marinum_M --------------------------------------------------
MUL_3865|M.ulcerans_Agy99 --------------------------------------------------
TH_4244|M.thermoresistible__bu --------------------------------------------------
MSMEG_2026|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0982|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
MAB_1305|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_4152|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIVLVHGFPDSHVLWD
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4002|M.marinum_M --------------------------------------------------
MUL_3865|M.ulcerans_Agy99 --------------------------------------------------
TH_4244|M.thermoresistible__bu --------------------------------------------------
MSMEG_2026|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0982|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
MAB_1305|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_4152|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVVPLLAKRFRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDELSP
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4002|M.marinum_M --------------------------------------------------
MUL_3865|M.ulcerans_Agy99 --------------------------------------------------
TH_4244|M.thermoresistible__bu --------------------------------------------------
MSMEG_2026|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0982|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
MAB_1305|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_4152|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSG
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4002|M.marinum_M --------------------------------------------------
MUL_3865|M.ulcerans_Agy99 --------------------------------------------------
TH_4244|M.thermoresistible__bu --------------------------------------------------
MSMEG_2026|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0982|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
MAB_1305|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_4152|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMV
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4002|M.marinum_M --------------------------------------------------
MUL_3865|M.ulcerans_Agy99 --------------------------------------------------
TH_4244|M.thermoresistible__bu --------------------------------------------------
MSMEG_2026|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0982|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
MAB_1305|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_4152|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQ
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4002|M.marinum_M --------------------------------------------------
MUL_3865|M.ulcerans_Agy99 --------------------------------------------------
TH_4244|M.thermoresistible__bu --------------------------------------------------
MSMEG_2026|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0982|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
MAB_1305|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_4152|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAAVHD
Mb1079|M.bovis_AF2122/97 --------------------MARQRFRDQVVLITGASSGIGEATAKAFAR
Rv1050|M.tuberculosis_H37Rv --------------------MARQRFRDQVVLITGASSGIGEATAKAFAR
MMAR_4002|M.marinum_M -----------------------MSCTRQHVVVTGASSGIGRSTALRLAS
MUL_3865|M.ulcerans_Agy99 ------------------------------MVVTGASSGIGRSTALRLAS
TH_4244|M.thermoresistible__bu --------------------------------VTGAARGIGRSIVTQLAD
MSMEG_2026|M.smegmatis_MC2_155 -------------------------MTT--WLITGCSTGIGRTLAEAVAA
Mvan_0982|M.vanbaalenii_PYR-1 --------------MPGHPTGIDPGMTEKVWFITGASRGFGREWAIAALD
MAV_2098|M.avium_104 -----------------------MTHRSPVVLVTGASSGIGRAVAKAFAA
MAB_1305|M.abscessus_ATCC_1997 ------------------------MSEHWTVLVTGASSGIGASAARIFAE
Mflv_4152|M.gilvum_PYR-GCK -------------------MAERAGAQRRVALITGASRGLGFASAVRLYR
MLBr_00862|M.leprae_Br4923 LADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAGSGIGRETALALAR
:**.. *:* .
Mb1079|M.bovis_AF2122/97 EGAVVALAARREGALRRVAREIEAAG---GRAMVAPLDVSSSESVRAMVA
Rv1050|M.tuberculosis_H37Rv EGAVVALAARREGALRRVAREIEAAG---GRAMVAPLDVSSSESVRAMVA
MMAR_4002|M.marinum_M AGWHVYAGVRRAADGEALQAASSGG-----QLSPLLMDVTVAEQIDAAVG
MUL_3865|M.ulcerans_Agy99 AGWHVYAGVRRAADGEALQAASSGG-----QLSPLLMDVTVAEQIDAAVG
TH_4244|M.thermoresistible__bu SGWDVVAGVRTEQDAENIVALNP-A-----RITSVLLDVTEADHIAALDR
MSMEG_2026|M.smegmatis_MC2_155 AGHNLVVTARDVSSVADIAETAP-D-----RVVAAPLDVTRPEQVTAAAQ
Mvan_0982|M.vanbaalenii_PYR-1 RGDKVAATARDLSTLDDLTAKYG-D-----ALLPLTLDVTDRDADFAAVT
MAV_2098|M.avium_104 KGFEVFGTTRNPRTAEPIPG-----------VELVELDVTDAASVDAAAK
MAB_1305|M.abscessus_ATCC_1997 RGHRVFGTSRNPDTITDRVPG----------VQYLRLDQTDKASIAECVA
Mflv_4152|M.gilvum_PYR-GCK EGWGVVAAMRTPERGMPRLREATGASEDDDRLVGVQLDLMDAASIAAAAK
MLBr_00862|M.leprae_Br4923 RGAEVVVSDIDEAAAKNTAAQIVASG---GVAYAYALDVSDAAAVEAFAE
* : :*
Mb1079|M.bovis_AF2122/97 DVVGEFG--RIDVVFNNAGVSLVGPVDAETFLDDTREMLEIDYLGTVRVV
Rv1050|M.tuberculosis_H37Rv DVVGEFG--RIDVVFNNAGVSLVGPVDAETFLDDTREMLEIDYLGTVRVV
MMAR_4002|M.marinum_M IVCEHIGAAGLDGLVDNAGIGVASPVEMVP-LDALRQQLEVNVVGQIAVT
MUL_3865|M.ulcerans_Agy99 IVGEHIGAAGLDGLVDNAGIGVASPVEMVP-LDALRQQLQVNVVGQIAVT
TH_4244|M.thermoresistible__bu SLDG-----RLDAVINNAGVVVPGPLETVS-PQDLRRQFDVNVIGHLAVT
MSMEG_2026|M.smegmatis_MC2_155 LADERFG--GVDVLVNNAGYGYRAAVEEGD-DADVRTLFDTHIFGTVATI
Mvan_0982|M.vanbaalenii_PYR-1 QAHDHFG--RLDIVVNNAGYGQFGFVEELS-EQEARAQIDTNVFGALWIT
MAV_2098|M.avium_104 SVIERTG--RIDILVNNAGSGILGAAEERS-VAQAQRLIDTNFLGLVRLT
MAB_1305|M.abscessus_ATCC_1997 QAGE------VDILVNNAGESQIGALEDVS-MDDFEKLYMTNVFGPVALT
Mflv_4152|M.gilvum_PYR-GCK SIDEAVG--APYAVVHNAGISAAGVVEETD-IELWERMFTTHVMGPVALT
MLBr_00862|M.leprae_Br4923 QVSAKHG--VPDIVVNNAGIGQAGRFLDTP-PEEFDRVLAVNLGGVVNCC
:..*** . . * :
Mb1079|M.bovis_AF2122/97 REVLPIMKQQRS-GRIMNMSSVVGRKAFARFAGYSSAMHAIAGFSDALRQ
Rv1050|M.tuberculosis_H37Rv REVLPIMKQQRS-GRIMNMSSVVGRKAFARFAGYSSAMHAIAGFSDALRQ
MMAR_4002|M.marinum_M QAFLPLLRRAA--GRIVVIASIGDRFTPPFGGPLAASKAAIATLADALRQ
MUL_3865|M.ulcerans_Agy99 QAFLPLLRRAA--GRIVVIASIGDRFTPPFGGPLAASKAAIATLADALRQ
TH_4244|M.thermoresistible__bu QAVLPRLRDSR--GRIVFISSLNGRISVPLMGAYCASKFALEAVADALRM
MSMEG_2026|M.smegmatis_MC2_155 KAVLPGMRARRR-GAIVNISSIGAQITPPGSGYYAAAKAAVEGLSGSLHT
Mvan_0982|M.vanbaalenii_PYR-1 QAALPYLRAQRS-GHLIQVSSIGGIVAFPNLGIYHASKWALEGFSQALAQ
MAV_2098|M.avium_104 NAVLPYLRAQGG-GRVINIGSVLGFLPQPYGALYAGSKHAVEGYSESLDH
MAB_1305|M.abscessus_ATCC_1997 KAVLPGMRERRR-GAVVMVGSMAGTLHVPFRSTYSSAKSALHTVADCLRY
Mflv_4152|M.gilvum_PYR-GCK KALLPSMRAAGQ-GRIVLVSSASGVRGQPATAPYSAAKGALERWGEAMAV
MLBr_00862|M.leprae_Br4923 RAFGQRLVERGTGGHIVNVSSMAAYAPLQSLSAYCTSKAATYMFSDCLRA
. : * :: :.* . : * . .:
Mb1079|M.bovis_AF2122/97 ELRGSGIAVSVIHPALTQTPLLAN--VDPADMPPPFRSLTPIPVHWVAAA
Rv1050|M.tuberculosis_H37Rv ELRGSGIAVSVIHPALTQTPLLAN--VDPADMPPPFRSLTPIPVHWVAAA
MMAR_4002|M.marinum_M EVAPWGIKVVMIEPASINSGAADKLERDAKKAIDEFGADGARLYRDAYLG
MUL_3865|M.ulcerans_Agy99 EEAPWGIKVVMIEPASINSGAADKLERDAKKAIDEFGADGARLYRDAYLG
TH_4244|M.thermoresistible__bu ELAPWRIPVIVIQPTQTDTDMVRGADVMVEQTAAAMRPEHRDLYARHIGG
MSMEG_2026|M.smegmatis_MC2_155 ELAPLGISVTVVEPGGFRTDFAGRSLKQSAAAIDDYADTAG-------KR
Mvan_0982|M.vanbaalenii_PYR-1 EVAEFGIHVTLIEPGGYSTDWAGASAKR-ATALPAYAEAHA-------AT
MAV_2098|M.avium_104 ETREFGVRVSVVEPAYTRTSFEAN-MADADAPIDAYAASRE-------HA
MAB_1305|M.abscessus_ATCC_1997 EVAPFGITVSTVEPGVIATGIETRRNRIVPDGSPYRDAFRT---------
Mflv_4152|M.gilvum_PYR-GCK EIAPFGLGVTILVTGTYDTDIITDAGTTDNRDLAGPYARIH-------QT
MLBr_00862|M.leprae_Br4923 ELDAVGVGLTTICPGVIDTNIIQSTRIDTPTAKQERVRDRR---------
* : : : . :
Mb1079|M.bovis_AF2122/97 VLDGVARRRARVVVPFQPRLLMVGDAFSPRYGDRVVRLLE----------
Rv1050|M.tuberculosis_H37Rv VLDGVARRRARVVVPFQPRLLMVGDAFSPRYGDRVVRLLE----------
MMAR_4002|M.marinum_M MVKAALSRERRGSPPTVVADLVLKVLTMARPHARNVVGKD----------
MUL_3865|M.ulcerans_Agy99 MVKAALSRERRGSPPTVVADLVLKVLTMARPHARNVGGKGRQDVGDRVPL
TH_4244|M.thermoresistible__bu MRKFIRQFHPSAVPADNVAAAVVNALTVRRPRSRYIVGITHQ--------
MSMEG_2026|M.smegmatis_MC2_155 RKEHDRVHGTQPGDPVKAAAAIIKAVESPEPPAFLLLGSD----------
Mvan_0982|M.vanbaalenii_PYR-1 ER-TRQARTARPGDPQASARALLTVVDAPEPPLRVFFGEL----------
MAV_2098|M.avium_104 RRRMAELMKT-ADDPAIVAEVVLKAATARKPKLRYPAGPL----------
MAB_1305|M.abscessus_ATCC_1997 VDAAMAAREEHGTATDELGRLVVDVALSPSPKPLYAKGSN----------
Mflv_4152|M.gilvum_PYR-GCK MDSRGRFAMGMARPPERFTDGLVKALEDTAAFRRHGVGPD----------
MLBr_00862|M.leprae_Br4923 -GQLDMMFRLRRYGPDKVADTILSAIKKNKPIRPVAPEAY----------
. ::
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4002|M.marinum_M --------------------------------------------------
MUL_3865|M.ulcerans_Agy99 ASDPGLGCAASQGFRASQAGFAGNIQVRVKRKVVDAMAPMEMPNQDQVQA
TH_4244|M.thermoresistible__bu --------------------------------------------------
MSMEG_2026|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0982|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
MAB_1305|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_4152|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00862|M.leprae_Br4923 --------------------------------------------------
Mb1079|M.bovis_AF2122/97 --------------------------------SKIFGRLIGSYRGS----
Rv1050|M.tuberculosis_H37Rv --------------------------------SKIFGRLIGSYRGS----
MMAR_4002|M.marinum_M --------------------------------ARMLATVSRWLPTP----
MUL_3865|M.ulcerans_Agy99 LDPVFGKMAVELGQHAWSLPQLTMRVKSFVFLAADLCTANLGFPLLTHVQ
TH_4244|M.thermoresistible__bu -------------------------------FPRLQAIILPNLPPA----
MSMEG_2026|M.smegmatis_MC2_155 --------------------------------ALSAYRRVAQARAE----
Mvan_0982|M.vanbaalenii_PYR-1 --------------------------------PLELARADYESRLA----
MAV_2098|M.avium_104 --------------------------------ARRLSLLKKFAPEA----
MAB_1305|M.abscessus_ATCC_1997 --------------------------------APIIMTAQRLLPAR----
Mflv_4152|M.gilvum_PYR-GCK --------------------------------ASMLLLSNRILPAAG---
MLBr_00862|M.leprae_Br4923 ----------------------------------ALYGISRVLPQVLR--
Mb1079|M.bovis_AF2122/97 ------------------------------VYRHQPTES---------AK
Rv1050|M.tuberculosis_H37Rv ------------------------------VYRHQPTES---------AK
MMAR_4002|M.marinum_M ------------------------------ALDALRRR----------VF
MUL_3865|M.ulcerans_Agy99 MAGFNGVTPAECAAGIRHLAPYVGYPTAAVALQQLRQQGPMGEEPAQAVE
TH_4244|M.thermoresistible__bu ------------------------------ARDPLLRR----------LG
MSMEG_2026|M.smegmatis_MC2_155 ------------------------------EIDKWADL----------TT
Mvan_0982|M.vanbaalenii_PYR-1 ------------------------------NWEKWQPV----------AI
MAV_2098|M.avium_104 ------------------------------VLDRGIRK----------TN
MAB_1305|M.abscessus_ATCC_1997 ------------------------------LFLKFLAR----------MH
Mflv_4152|M.gilvum_PYR-GCK ------------------------MHHMSRVVLGIPRQGAMRDGAWPLTT
MLBr_00862|M.leprae_Br4923 ------------------------------STARIRVI------------
Mb1079|M.bovis_AF2122/97 AQAAQPERGYSSAR------------------------------------
Rv1050|M.tuberculosis_H37Rv AQAAQPERGYSSAR------------------------------------
MMAR_4002|M.marinum_M GLPKPGSLAISRSA------------------------------------
MUL_3865|M.ulcerans_Agy99 PQPLPGHIGEALDALDKDFARFVTGQFHQRWAGDQRLTPRERALSCLAAD
TH_4244|M.thermoresistible__bu SQP-----------------------------------------------
MSMEG_2026|M.smegmatis_MC2_155 STDFDS--------------------------------------------
Mvan_0982|M.vanbaalenii_PYR-1 EAQG----------------------------------------------
MAV_2098|M.avium_104 GLPAASRPHSHLQL------------------------------------
MAB_1305|M.abscessus_ATCC_1997 GLKVR---------------------------------------------
Mflv_4152|M.gilvum_PYR-GCK AQRAMVAAAKVIPQPVMQRLVQWSTRRAAGSGGGTSEG------------
MLBr_00862|M.leprae_Br4923 --------------------------------------------------
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4002|M.marinum_M --------------------------------------------------
MUL_3865|M.ulcerans_Agy99 VLNQTLDESFAMHVELALAAGAGPAQLEAVLLLVAEYGIPKAWRAYRALD
TH_4244|M.thermoresistible__bu --------------------------------------------------
MSMEG_2026|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0982|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
MAB_1305|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_4152|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00862|M.leprae_Br4923 --------------------------------------------------
Mb1079|M.bovis_AF2122/97 ----
Rv1050|M.tuberculosis_H37Rv ----
MMAR_4002|M.marinum_M ----
MUL_3865|M.ulcerans_Agy99 ALTA
TH_4244|M.thermoresistible__bu ----
MSMEG_2026|M.smegmatis_MC2_155 ----
Mvan_0982|M.vanbaalenii_PYR-1 ----
MAV_2098|M.avium_104 ----
MAB_1305|M.abscessus_ATCC_1997 ----
Mflv_4152|M.gilvum_PYR-GCK ----
MLBr_00862|M.leprae_Br4923 ----