For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVTGASSGIGRSTALRLASAGWHVYAGVRRAADGEALQAASSGGQLSPLLMDVTVAEQIDAAVGIVGEHI GAAGLDGLVDNAGIGVASPVEMVPLDALRQQLQVNVVGQIAVTQAFLPLLRRAAGRIVVIASIGDRFTPP FGGPLAASKAAIATLADALRQEEAPWGIKVVMIEPASINSGAADKLERDAKKAIDEFGADGARLYRDAYL GMVKAALSRERRGSPPTVVADLVLKVLTMARPHARNVGGKGRQDVGDRVPLASDPGLGCAASQGFRASQA GFAGNIQVRVKRKVVDAMAPMEMPNQDQVQALDPVFGKMAVELGQHAWSLPQLTMRVKSFVFLAADLCTA NLGFPLLTHVQMAGFNGVTPAECAAGIRHLAPYVGYPTAAVALQQLRQQGPMGEEPAQAVEPQPLPGHIG EALDALDKDFARFVTGQFHQRWAGDQRLTPRERALSCLAADVLNQTLDESFAMHVELALAAGAGPAQLEA VLLLVAEYGIPKAWRAYRALDALTA*
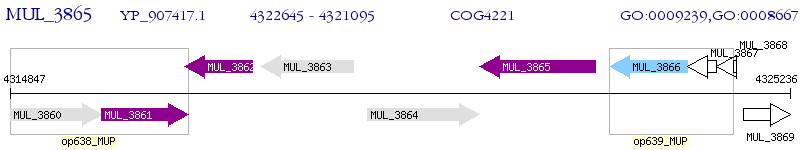
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_3865 | - | - | 100% (516) | dehydrogenase/decarboxylase protein |
| M. ulcerans Agy99 | MUL_3423 | - | 9e-24 | 31.28% (243) | short chain dehydrogenase |
| M. ulcerans Agy99 | MUL_2794 | - | 1e-19 | 32.34% (235) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1079 | - | 5e-17 | 34.97% (183) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_0295 | - | 4e-22 | 33.33% (255) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv1050 | - | 5e-17 | 34.97% (183) | oxidoreductase |
| M. leprae Br4923 | MLBr_01740 | - | 1e-14 | 27.89% (190) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1305 | - | 1e-21 | 33.20% (244) | short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_4002 | - | 1e-133 | 98.41% (251) | dehydrogenase/decarboxylase protein |
| M. avium 104 | MAV_2098 | - | 2e-23 | 32.27% (251) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2026 | - | 6e-24 | 36.18% (199) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_4244 | - | 3e-37 | 38.89% (252) | PUTATIVE putative Dehydrogenase/decarboxylase protein |
| M. vanbaalenii PYR-1 | Mvan_0982 | - | 1e-20 | 33.47% (245) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1079|M.bovis_AF2122/97 ------MARQRFRDQVVLITGASSGIGEATAKAFAREGAVVALAARREGA
Rv1050|M.tuberculosis_H37Rv ------MARQRFRDQVVLITGASSGIGEATAKAFAREGAVVALAARREGA
MUL_3865|M.ulcerans_Agy99 ----------------MVVTGASSGIGRSTALRLASAGWHVYAGVRRAAD
MMAR_4002|M.marinum_M ---------MSCTRQHVVVTGASSGIGRSTALRLASAGWHVYAGVRRAAD
TH_4244|M.thermoresistible__bu ------------------VTGAARGIGRSIVTQLADSGWDVVAGVRTEQD
MAB_1305|M.abscessus_ATCC_1997 ----------MSEHWTVLVTGASSGIGASAARIFAERGHRVFGTSRNPDT
MAV_2098|M.avium_104 ---------MTHRSPVVLVTGASSGIGRAVAKAFAAKGFEVFGTTRNP-R
Mflv_0295|M.gilvum_PYR-GCK ------------MSKVLLISGASRGLGRAITESALAAGHLVVAGVRATSS
MSMEG_2026|M.smegmatis_MC2_155 -------------MTTWLITGCSTGIGRTLAEAVAAAGHNLVVTARDVSS
Mvan_0982|M.vanbaalenii_PYR-1 MPGHPTGIDPGMTEKVWFITGASRGFGREWAIAALDRGDKVAATARDLST
MLBr_01740|M.leprae_Br4923 ----MVKTAQYFVGKRCFVTGAASGIGRATALRLAAYGAELYLTDRHCDG
::*.: *:* . * : *
Mb1079|M.bovis_AF2122/97 LRRVAREIEAA-GGRAMVAPLDVSSSESVRAMVADVVGEFG--RIDVVFN
Rv1050|M.tuberculosis_H37Rv LRRVAREIEAA-GGRAMVAPLDVSSSESVRAMVADVVGEFG--RIDVVFN
MUL_3865|M.ulcerans_Agy99 GEALQAASSG---GQLSPLLMDVTVAEQIDAAVGIVGEHIGAAGLDGLVD
MMAR_4002|M.marinum_M GEALQAASSG---GQLSPLLMDVTVAEQIDAAVGIVCEHIGAAGLDGLVD
TH_4244|M.thermoresistible__bu AENIVALNP----ARITSVLLDVTEADHIAALDRSLDG-----RLDAVIN
MAB_1305|M.abscessus_ATCC_1997 ITDRVPG--------VQYLRLDQTDKASIAECVA----QAG--EVDILVN
MAV_2098|M.avium_104 TAEPIPG--------VELVELDVTDAASVDAAAKSVIERTG--RIDILVN
Mflv_0295|M.gilvum_PYR-GCK LSDLATREP----DRLAVVELDVTDDRHARAAVATAVERFG--RLDVLVN
MSMEG_2026|M.smegmatis_MC2_155 VADIAETAP----DRVVAAPLDVTRPEQVTAAAQLADERFG--GVDVLVN
Mvan_0982|M.vanbaalenii_PYR-1 LDDLTAKYG----DALLPLTLDVTDRDADFAAVTQAHDHFG--RLDIVVN
MLBr_01740|M.leprae_Br4923 LAQTVSEARALGAQVPEYRALDISDYDEVAAFGADIHARHP--SMDIVMN
:* : :* :.:
Mb1079|M.bovis_AF2122/97 NAGVSLVGPVDAETFLDDTREMLEIDYLGTVRVVREVLP-IMKQQRSGRI
Rv1050|M.tuberculosis_H37Rv NAGVSLVGPVDAETFLDDTREMLEIDYLGTVRVVREVLP-IMKQQRSGRI
MUL_3865|M.ulcerans_Agy99 NAGIGVASPVEMVP-LDALRQQLQVNVVGQIAVTQAFLP-LLR-RAAGRI
MMAR_4002|M.marinum_M NAGIGVASPVEMVP-LDALRQQLEVNVVGQIAVTQAFLP-LLR-RAAGRI
TH_4244|M.thermoresistible__bu NAGVVVPGPLETVS-PQDLRRQFDVNVIGHLAVTQAVLP-RLR-DSRGRI
MAB_1305|M.abscessus_ATCC_1997 NAGESQIGALEDVS-MDDFEKLYMTNVFGPVALTKAVLP-GMRERRRGAV
MAV_2098|M.avium_104 NAGSGILGAAEERS-VAQAQRLIDTNFLGLVRLTNAVLP-YLRAQGGGRV
Mflv_0295|M.gilvum_PYR-GCK NAGYANMSAIEDVD-VDDFRAQIETNFFGVVNLTRAALP-AMRRQRDGHI
MSMEG_2026|M.smegmatis_MC2_155 NAGYGYRAAVEEGD-DADVRTLFDTHIFGTVATIKAVLP-GMRARRRGAI
Mvan_0982|M.vanbaalenii_PYR-1 NAGYGQFGFVEELS-EQEARAQIDTNVFGALWITQAALP-YLRAQRSGHL
MLBr_01740|M.leprae_Br4923 IAGVSVWGTVDRLT-HDQWSNVVAINLMGPIHIIETFVPAILAAGRGGHL
** . : . .* : . :* : * :
Mb1079|M.bovis_AF2122/97 MNMSSVVGRKAFARFAGYSSAMHAIAGFSDALRQELRGSGIAVSVIHPAL
Rv1050|M.tuberculosis_H37Rv MNMSSVVGRKAFARFAGYSSAMHAIAGFSDALRQELRGSGIAVSVIHPAL
MUL_3865|M.ulcerans_Agy99 VVIASIGDRFTPPFGGPLAASKAAIATLADALRQEEAPWGIKVVMIEPAS
MMAR_4002|M.marinum_M VVIASIGDRFTPPFGGPLAASKAAIATLADALRQEVAPWGIKVVMIEPAS
TH_4244|M.thermoresistible__bu VFISSLNGRISVPLMGAYCASKFALEAVADALRMELAPWRIPVIVIQPTQ
MAB_1305|M.abscessus_ATCC_1997 VMVGSMAGTLHVPFRSTYSSAKSALHTVADCLRYEVAPFGITVSTVEPGV
MAV_2098|M.avium_104 INIGSVLGFLPQPYGALYAGSKHAVEGYSESLDHETREFGVRVSVVEPAY
Mflv_0295|M.gilvum_PYR-GCK VQISSVGGRLVRPGLAAYQSAKWAVTGFSGVLAVEVAPLGIKVTVLEPGG
MSMEG_2026|M.smegmatis_MC2_155 VNISSIGAQITPPGSGYYAAAKAAVEGLSGSLHTELAPLGISVTVVEPGG
Mvan_0982|M.vanbaalenii_PYR-1 IQVSSIGGIVAFPNLGIYHASKWALEGFSQALAQEVAEFGIHVTLIEPGG
MLBr_01740|M.leprae_Br4923 VNVSSAAGLVALPWHAAYSASKYGLRGLSEVLRFDLARYRIGVSVVVPGA
: :.* . . .: .: : * : : * : *
Mb1079|M.bovis_AF2122/97 TQTPLLANVDP--ADMPPPFRSLTPIPVHWVAAAVLDGVARRRA--RVVV
Rv1050|M.tuberculosis_H37Rv TQTPLLANVDP--ADMPPPFRSLTPIPVHWVAAAVLDGVARRRA--RVVV
MUL_3865|M.ulcerans_Agy99 INSGAADKLERDAKKAIDEFGADGARLYRDAYLGMVKAALSRER--RGSP
MMAR_4002|M.marinum_M INSGAADKLERDAKKAIDEFGADGARLYRDAYLGMVKAALSRER--RGSP
TH_4244|M.thermoresistible__bu TDTDMVRGADVMVEQTAAAMRPEHRDLYARHIGGMRKFIRQFHP--SAVP
MAB_1305|M.abscessus_ATCC_1997 IATGIETRRNR-IVPDGSPYRDAFRTVDAAMAAREEHGT----------A
MAV_2098|M.avium_104 TRTSFEANMAD-ADAPIDAYAASREHARRRMAELMKTAD----------D
Mflv_0295|M.gilvum_PYR-GCK MRTDWAGSSMR-IAPVREEYAATVGAAASLSDPNVLGAS----------D
MSMEG_2026|M.smegmatis_MC2_155 FRTDFAGRSLKQSAAAIDDYADTAGKRRKEHDR-VHGTQPG--------D
Mvan_0982|M.vanbaalenii_PYR-1 YSTDWAGASAK-RATALPAYAEAH--AATERTRQARTARPG--------D
MLBr_01740|M.leprae_Br4923 VRTPLVDTIEIAGVDRQDPRFSRWVDRFRGHAVSAERAAEKILAGVAKNR
:
Mb1079|M.bovis_AF2122/97 PFQPRLLMVGDAFSPRYGDRVVRLLE------------------------
Rv1050|M.tuberculosis_H37Rv PFQPRLLMVGDAFSPRYGDRVVRLLE------------------------
MUL_3865|M.ulcerans_Agy99 PTVVADLVLKVLTMARPHARNVGGKGRQDVGDRVPLASDPGLGCAASQGF
MMAR_4002|M.marinum_M PTVVADLVLKVLTMARPHARNVVGKD------------------------
TH_4244|M.thermoresistible__bu ADNVAAAVVNALTVRRPRSRYIVGITHQ----------------------
MAB_1305|M.abscessus_ATCC_1997 TDELGRLVVDVALSPSPKPLYAKGSN------------------------
MAV_2098|M.avium_104 PAIVAEVVLKAATARKPKLRYPAGPL------------------------
Mflv_0295|M.gilvum_PYR-GCK PAKVAGLLLDVIGMPDPPRRLLVGPD------------------------
MSMEG_2026|M.smegmatis_MC2_155 PVKAAAAIIKAVESPEPPAFLLLGSD------------------------
Mvan_0982|M.vanbaalenii_PYR-1 PQASARALLTVVDAPEPPLRVFFGEL------------------------
MLBr_01740|M.leprae_Br4923 YLIYTSHDIRVLYGFKRLAWWPYGVVMK----------------------
:
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
MUL_3865|M.ulcerans_Agy99 RASQAGFAGNIQVRVKRKVVDAMAPMEMPNQDQVQALDPVFGKMAVELGQ
MMAR_4002|M.marinum_M --------------------------------------------------
TH_4244|M.thermoresistible__bu --------------------------------------------------
MAB_1305|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
Mflv_0295|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_2026|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0982|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01740|M.leprae_Br4923 --------------------------------------------------
Mb1079|M.bovis_AF2122/97 ------------------SKIFGRLIGSYR--------------------
Rv1050|M.tuberculosis_H37Rv ------------------SKIFGRLIGSYR--------------------
MUL_3865|M.ulcerans_Agy99 HAWSLPQLTMRVKSFVFLAADLCTANLGFPLLTHVQMAGFNGVTPAECAA
MMAR_4002|M.marinum_M ------------------ARMLATVSRWLP-------------TP-----
TH_4244|M.thermoresistible__bu -----------------FPRLQAIILPNLP--------------P-----
MAB_1305|M.abscessus_ATCC_1997 ------------------APIIMTAQRLLP--------------------
MAV_2098|M.avium_104 ------------------ARRLSLLKKFAP--------------------
Mflv_0295|M.gilvum_PYR-GCK ------------------AYRYATAAGRDL--------------------
MSMEG_2026|M.smegmatis_MC2_155 ------------------ALSAYRRVAQAR--------------------
Mvan_0982|M.vanbaalenii_PYR-1 ------------------PLELARADYESR--------------------
MLBr_01740|M.leprae_Br4923 ----------------QVNVVFTRALRPSP--------------------
Mb1079|M.bovis_AF2122/97 --------------GSVYRHQPTES---------AKAQAAQPERGYSSAR
Rv1050|M.tuberculosis_H37Rv --------------GSVYRHQPTES---------AKAQAAQPERGYSSAR
MUL_3865|M.ulcerans_Agy99 GIRHLAPYVGYPTAAVALQQLRQQGPMGEEPAQAVEPQPLPGHIGEALDA
MMAR_4002|M.marinum_M ----------------ALDALRRR----------VFGLPKPGSLAISRSA
TH_4244|M.thermoresistible__bu ---------------AARDPLLRR----------LGSQP-----------
MAB_1305|M.abscessus_ATCC_1997 --------------ARLFLKFLAR----------MHGLKVR---------
MAV_2098|M.avium_104 --------------EAVLDRGIRK----------TNGLPAASRPHSHLQL
Mflv_0295|M.gilvum_PYR-GCK --------------LAVDERWRTL----------SMSTAADDVSPEQLDP
MSMEG_2026|M.smegmatis_MC2_155 --------------AEEIDKWADL----------TTSTDFDS--------
Mvan_0982|M.vanbaalenii_PYR-1 --------------LANWEKWQPV----------AIEAQG----------
MLBr_01740|M.leprae_Br4923 ---------------ATMRCVEPIKVG-------VYSQPRSGEGGKSGN-
Mb1079|M.bovis_AF2122/97 --------------------------------------------------
Rv1050|M.tuberculosis_H37Rv --------------------------------------------------
MUL_3865|M.ulcerans_Agy99 LDKDFARFVTGQFHQRWAGDQRLTPRERALSCLAADVLNQTLDESFAMHV
MMAR_4002|M.marinum_M --------------------------------------------------
TH_4244|M.thermoresistible__bu --------------------------------------------------
MAB_1305|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
Mflv_0295|M.gilvum_PYR-GCK LGSASR--------------------------------------------
MSMEG_2026|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0982|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01740|M.leprae_Br4923 --------------------------------------------------
Mb1079|M.bovis_AF2122/97 ----------------------------------------
Rv1050|M.tuberculosis_H37Rv ----------------------------------------
MUL_3865|M.ulcerans_Agy99 ELALAAGAGPAQLEAVLLLVAEYGIPKAWRAYRALDALTA
MMAR_4002|M.marinum_M ----------------------------------------
TH_4244|M.thermoresistible__bu ----------------------------------------
MAB_1305|M.abscessus_ATCC_1997 ----------------------------------------
MAV_2098|M.avium_104 ----------------------------------------
Mflv_0295|M.gilvum_PYR-GCK ----------------------------------------
MSMEG_2026|M.smegmatis_MC2_155 ----------------------------------------
Mvan_0982|M.vanbaalenii_PYR-1 ----------------------------------------
MLBr_01740|M.leprae_Br4923 ----------------------------------------