For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRIIDADGHVAENASLTGEALRRWPDRVRLTTRGRPQLMIEGRHYPQDNGPGAGCPPQHGISKAPGINCA TADGVLADADRDHLDTMVLYPSVGLCVPSLEDPAFAAGFARLYNEWIADFCAPTNGRLRGVGVTPIEHGQ VAIDIMKEANELGLVATLVPPALKSRNLDHADLDPFYAAAVEQGMPLGIHGAPGMHLPKIGVDRFTNYIQ VHCISFPFDQMTAMTALVSGGVFDRHPRLRVAFLEAGVGWVPFFVDRLHEHYEKRGDWIDGGWRRDPHDY LRAGNVWVTCEPDEPILPGVIDVLGDDFIMFASDYPHWDGEWPESTKQLRTRTDIGEQSRNKIAGLNAQR FYELN
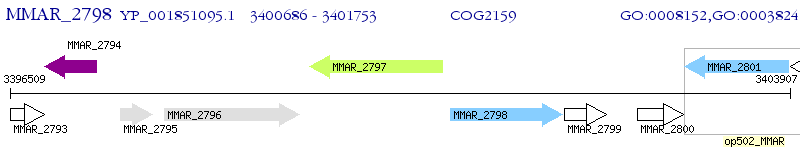
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2798 | - | - | 100% (355) | amidohydrolase |
| M. marinum M | MMAR_4742 | - | 3e-16 | 22.61% (314) | metal-dependent amidohydrolase |
| M. marinum M | MMAR_4820 | - | 2e-15 | 27.24% (257) | hypothetical protein MMAR_4820 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2419 | - | 2e-16 | 25.35% (288) | amidohydrolase 2 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_02013 | - | 5e-39 | 61.21% (116) | hypothetical protein MLBr_02013 |
| M. abscessus ATCC 19977 | MAB_0299 | - | 2e-11 | 24.43% (262) | putative amidohydrolase (aminocarboxymuconate-semialdehyde |
| M. avium 104 | MAV_2798 | - | 0.0 | 81.13% (355) | amidohydrolase family protein |
| M. smegmatis MC2 155 | MSMEG_4837 | - | 1e-16 | 24.28% (276) | amidohydrolase 2 |
| M. thermoresistible (build 8) | TH_2827 | - | 1e-16 | 26.36% (368) | PUTATIVE amidohydrolase 2 |
| M. ulcerans Agy99 | MUL_0389 | - | 4e-16 | 27.63% (257) | hypothetical protein MUL_0389 |
| M. vanbaalenii PYR-1 | Mvan_0901 | - | 5e-18 | 24.07% (295) | amidohydrolase 2 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2798|M.marinum_M -----------------------------------------MRIIDADGH
MAV_2798|M.avium_104 -----------------------------------------MRIIDADGH
MLBr_02013|M.leprae_Br4923 --------------------------------------------------
TH_2827|M.thermoresistible__bu ------------------------------------VTAPALEIWDADNH
Mvan_0901|M.vanbaalenii_PYR-1 -------------------------------------MGSSTLIIDTDTH
MUL_0389|M.ulcerans_Agy99 --------------------------------------MDRYTVISADCH
MSMEG_4837|M.smegmatis_MC2_155 ------------------------------------MNKDDMILISVDDH
Mflv_2419|M.gilvum_PYR-GCK MSTPTVPSPAHPPTLYPPEGFGAPKHRHGHSTGAVTGLPADTVIFSADNH
MAB_0299|M.abscessus_ATCC_1997 -----------------------------------------MTSVRIDIH
MMAR_2798|M.marinum_M VAENASLTGEALRRWP----------DRVRLTTRGRPQLMIEGRHYPQDN
MAV_2798|M.avium_104 VAENSSLAIEAIQRWP----------EHVKPGRHGRLGLMIEGRNYPEDG
MLBr_02013|M.leprae_Br4923 --------------------------------------------------
TH_2827|M.thermoresistible__bu LYEPTDAYTRHLPKQYRGAIQWIQVDGLTKLMIKGRLTDTIPNPTYEVVA
Mvan_0901|M.vanbaalenii_PYR-1 ITEPPDLWTSRMSKSRWG-----NLIPEVKWVEEKQGEYWCMDGQPVFTV
MUL_0389|M.ulcerans_Agy99 AGADLFDYREYLEAKY-------------------QDEFDDWAKTFVNPF
MSMEG_4837|M.smegmatis_MC2_155 IIEPPDMFKNHLPAKYR------DEAPRLVHNPDGSDTWQFRDTVIPNVA
Mflv_2419|M.gilvum_PYR-GCK ISVADDIFYERFPEELKG----------------AAPRIWYEDGAYMVGM
MAB_0299|M.abscessus_ATCC_1997 AHLWSDEYLSLLDGYGR------------------------PATDVHRGL
MMAR_2798|M.marinum_M GPGAGCPPQHG-ISKAPG------------INCATADGVLADADRDHLDT
MAV_2798|M.avium_104 GPGAGCPPEHG-ISDAPD------------INCRSAEGVLGDADRDHIDT
MLBr_02013|M.leprae_Br4923 --------------------------------------MLRDVDREHIDM
TH_2827|M.thermoresistible__bu SPGAWADYFRG-INPEGKSLRELANPIRCPEEFRHAEQRLRLLDAQGIQA
Mvan_0901|M.vanbaalenii_PYR-1 GTCIMVPGEDGKPVRSPRFPDYADRFSSMHPSAYDAKARLEVMDAYGIQA
MUL_0389|M.ulcerans_Agy99 GDLSEPDAEHN----------------------WDSDRRNAELDSDGVAG
MSMEG_4837|M.smegmatis_MC2_155 LNAVAGRPKEEYGLEPQG-------LDEIRKGCYDPNERVKDMNAGGVLA
Mflv_2419|M.gilvum_PYR-GCK KGKAWTGGDFGRVLMQYDDLAG--------AASNNIEARIRELKEDGIDK
MAB_0299|M.abscessus_ATCC_1997 GAGATRDDLDG---------------------------RFELMDEAGVDL
. :
MMAR_2798|M.marinum_M MVLYPSVG-----------LCVPSLEDPAFAAGFARLYNEWIADFCAPT-
MAV_2798|M.avium_104 MVLYPSLG-----------LCAPSLRDPEFAAGFARLYNRWIADYCAQS-
MLBr_02013|M.leprae_Br4923 MVLYPSLG-----------FCILRLDDPDFATRLARFYNQWIGDYCAPT-
TH_2827|M.thermoresistible__bu CVMFPTTGGM---------LEERMLDDIELTHAVVHAYNEWLLEEWTFDY
Mvan_0901|M.vanbaalenii_PYR-1 AAIFPNLGFVGP-----NIFAAAGPDALEFQTAALQAYNDFLLDWSSIA-
MUL_0389|M.ulcerans_Agy99 EVIFPNTVPPFFPQGSLAAPAPSTARDLELRWAGLRAHNRWLADFCSLS-
MSMEG_4837|M.smegmatis_MC2_155 TMNFPSFPG--------FAARLFATDDEEFSLALVQAYNDWHIDEWCASN
Mflv_2419|M.gilvum_PYR-GCK ELAFPNAV-----------LALFHYPDKALRERVFRIYNEHIADLQERS-
MAB_0299|M.abscessus_ATCC_1997 QILTATPAS-------------PHFEDQAAAVASARFINDEYAKLSSEY-
.. : * .
MMAR_2798|M.marinum_M NGRLRGVGVTPIEHGQVAIDIMK-EANELGLVATLVPP------ALKSRN
MAV_2798|M.avium_104 GGRLRGVAVTPIEHGQLAIDVMR-EAKDLGLVATLVPP------ALQSRN
MLBr_02013|M.leprae_Br4923 NGWLRGGGVTSMERGQVAIDITN-GVKELGIAVTLIPP------VLNASN
TH_2827|M.thermoresistible__bu AGRIFATPVITLPDVDKAVAELN-WCVENGARAVLVNPRPVATITGHTTS
Mvan_0901|M.vanbaalenii_PYR-1 PDRLLSLALIPYWDVDAAVAEIE-RCAAAGHKGLVSTG---KPHEHGYKL
MUL_0389|M.ulcerans_Agy99 PERRAGVGQILLGDVDEAVAEVA-QIAKLGLPGGVLLPG--VAPGTGIPA
MSMEG_4837|M.smegmatis_MC2_155 PGRFIPMAIPAIWNPGLAAAEVR-RVAEKGVHSLTFTEN--PSTLGYPSF
Mflv_2419|M.gilvum_PYR-GCK NGHFYGVGLINWWDPKGTRSTLE-ELKSLGLKTFLLPLNPGKDDEGNIYD
MAB_0299|M.abscessus_ATCC_1997 PGRFGALASLPLPHVPAALEELARAIDELGMYGASITTS------VLGRS
: *
MMAR_2798|M.marinum_M LDHADLDPFYAAAVEQGMPLGIHGAPGMHLPKIGVDRFTNYIQVHCISFP
MAV_2798|M.avium_104 LDHPDLDPFYAAAVELGMPLGVHGAPGIHLPKIGVDRFTNYIQVHCISFP
MLBr_02013|M.leprae_Br4923 LDHPYLGPFYAATVERGMAISIHAR-----------------------YP
TH_2827|M.thermoresistible__bu MGQPYFDPFWHRVEQLGIPVLMHACDSGYDRYSRDWEGKGGEYTPFKPDA
Mvan_0901|M.vanbaalenii_PYR-1 LADRYWDPMWAAAQDANLSISFHVGGGNLSRHLNDERTSVEGWRATLARL
MUL_0389|M.ulcerans_Agy99 LYAEHWEPLWAACAQAGLVINHHGGNAGPSPIDGWGSSLAVLVYESHWWA
MSMEG_4837|M.smegmatis_MC2_155 HDLEYWRPLWQALVDTETVMNVHIGSSGKLAITAPDAPMDVMITLQPMNI
Mflv_2419|M.gilvum_PYR-GCK YGSTSMDAVWDEIEAAGLPVSHHIGETPPKTPCQNNSVVVGMMVNVDSFR
MAB_0299|M.abscessus_ATCC_1997 IADPIFDPLYAELNRRRAVLFVHPAGCGAESSLITDHSLTWSIGAPIEDT
..: : *
MMAR_2798|M.marinum_M FDQMTAMTA---------LVSGGVFDRHPRLRVAFLEAGVGWVPFFVDRL
MAV_2798|M.avium_104 FDQMTAMTA---------MVSGGVFERHPKLRVAFLEAGAGWVPFFIDRL
MLBr_02013|M.leprae_Br4923 F-------------------------------------AADWC-------
TH_2827|M.thermoresistible__bu FRTIVYEDARSILDTCAALVAHGVFTRHPGVRVGVVENGGAWVPRLIDLF
Mvan_0901|M.vanbaalenii_PYR-1 TTSFFLESG----ITLADLLMSGVLARYPKLRFVSVESAVGWIPFLLESL
MUL_0389|M.ulcerans_Agy99 HRALLH------------LIFSGVLDRYPDLTVVLTEQGTGWIPATLDSL
MSMEG_4837|M.smegmatis_MC2_155 VQAAAD------------LLWSAPIKEFPTLKIALSEGGTGWIPYFLDRV
Mflv_2419|M.gilvum_PYR-GCK EQFAKY-------------VFSGILDRHPSLKIGWFEGGIAWVPTALQDA
MAB_0299|M.abscessus_ATCC_1997 VAIMHL-------------IVAGVPSRYPDMKIVTCHLG-GALPMVLERA
.
MMAR_2798|M.marinum_M HEHYEKR-------GDWIDGG--WRRDPHDYLRAGNVWVTCEPDEPILPG
MAV_2798|M.avium_104 HEHYEKR-------GDWVERG--WRRDPHEYLKAGNIFVTCEPEEPILPG
MLBr_02013|M.leprae_Br4923 --------------------------------------------------
TH_2827|M.thermoresistible__bu ERVYKKM-------PNEFGAH--PVEQFREHVWVN------PFHEEDLSG
Mvan_0901|M.vanbaalenii_PYR-1 DFHYKKY-------EPWLERPEFAGNDMLPSDYFHRQVYANYWYEDLQPW
MUL_0389|M.ulcerans_Agy99 DVAAARYRRPNSAIARFAGPTAGSLPLQPSQYWARQCYVGASFLRPVECA
MSMEG_4837|M.smegmatis_MC2_155 DRTFEMH-------STWTHQN---FGGKLPSEVFREHFMTCFISDPVGVK
Mflv_2419|M.gilvum_PYR-GCK EHMLASY-----------RHMFNHELQHDVNHYWANHMSASFMVDPLGLQ
MAB_0299|M.abscessus_ATCC_1997 HRQVTEW------------EATQCPEAPRDAARRLWYDTVGHDHTPALRA
MMAR_2798|M.marinum_M VIDVLGDDFIMFASDYPHWDGEWPESTKQLR---TRTDIGEQSRNKIAGL
MAV_2798|M.avium_104 VIDVLGADFIMFASDYPHWDGEWPESTKHLR---TRADISEETREKIGGL
MLBr_02013|M.leprae_Br4923 --------------------------------------------------
TH_2827|M.thermoresistible__bu LIDLIGADRVLFGSDFPHPEG-LAEPAKLTP---EIAELPEPTIEAVMGG
Mvan_0901|M.vanbaalenii_PYR-1 HVAAIGEDNLMFETDYPHQTCLDKEEIQHSIN-EGLSGVSETTKEKILWR
MUL_0389|M.ulcerans_Agy99 QRHRIGVDRIMWASDYPHMEGTAPYSREALR--HTFSDVDPDEVAAMVGG
MSMEG_4837|M.smegmatis_MC2_155 NRHMIGIDNICWEMDYPHSDSMWPGAPEELSAVFDTYDVPDDEINKITHE
Mflv_2419|M.gilvum_PYR-GCK LIDRIGVDNVMWSSDYPHNESTFGYSEKSLAT--VVDAVGPEAAVKIVSS
MAB_0299|M.abscessus_ATCC_1997 AVASLGVDRLIFGSDFPYQAGPRYVSSATYIE----EGLSAEDASAILDH
MMAR_2798|M.marinum_M NAQRFYELN-----------------------------------------
MAV_2798|M.avium_104 NAQRFYNLN-----------------------------------------
MLBr_02013|M.leprae_Br4923 --------------------------------------------------
TH_2827|M.thermoresistible__bu NLRELLTAVPHGR-------------------------------------
Mvan_0901|M.vanbaalenii_PYR-1 NAASLFNMDIAKLEKLT---------------------------------
MUL_0389|M.ulcerans_Agy99 NAAAVYGFDLQALAPLAARIGPTVTEVAEPLAAIPADASSTAFEPDPIRA
MSMEG_4837|M.smegmatis_MC2_155 NAMRLYHFDPFAHVPKEQATVGALRKAAEGHDVSIRALSKKDKTGASFAD
Mflv_2419|M.gilvum_PYR-GCK NVQRFLGL------------------------------------------
MAB_0299|M.abscessus_ATCC_1997 NGAELLGLTGV---------------------------------------
MMAR_2798|M.marinum_M -------------
MAV_2798|M.avium_104 -------------
MLBr_02013|M.leprae_Br4923 -------------
TH_2827|M.thermoresistible__bu -------------
Mvan_0901|M.vanbaalenii_PYR-1 -------------
MUL_0389|M.ulcerans_Agy99 W------------
MSMEG_4837|M.smegmatis_MC2_155 FQANAKAVSGARD
Mflv_2419|M.gilvum_PYR-GCK -------------
MAB_0299|M.abscessus_ATCC_1997 -------------