For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NKDDMILISVDDHIIEPPDMFKNHLPAKYRDEAPRLVHNPDGSDTWQFRDTVIPNVALNAVAGRPKEEYG LEPQGLDEIRKGCYDPNERVKDMNAGGVLATMNFPSFPGFAARLFATDDEEFSLALVQAYNDWHIDEWCA SNPGRFIPMAIPAIWNPGLAAAEVRRVAEKGVHSLTFTENPSTLGYPSFHDLEYWRPLWQALVDTETVMN VHIGSSGKLAITAPDAPMDVMITLQPMNIVQAAADLLWSAPIKEFPTLKIALSEGGTGWIPYFLDRVDRT FEMHSTWTHQNFGGKLPSEVFREHFMTCFISDPVGVKNRHMIGIDNICWEMDYPHSDSMWPGAPEELSAV FDTYDVPDDEINKITHENAMRLYHFDPFAHVPKEQATVGALRKAAEGHDVSIRALSKKDKTGASFADFQA NAKAVSGARD*
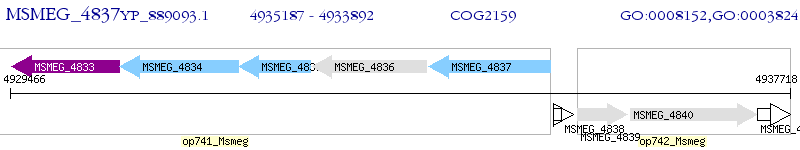
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4837 | - | - | 100% (431) | amidohydrolase 2 |
| M. smegmatis MC2 155 | MSMEG_6547 | - | e-116 | 49.38% (405) | amidohydrolase 2 |
| M. smegmatis MC2 155 | MSMEG_4855 | - | 8e-17 | 22.07% (358) | amidohydrolase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2441 | - | 0.0 | 78.65% (431) | amidohydrolase 2 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4742 | - | 0.0 | 79.21% (433) | metal-dependent amidohydrolase |
| M. avium 104 | MAV_0907 | - | 0.0 | 78.98% (433) | amidohydrolase 2 |
| M. thermoresistible (build 8) | TH_2339 | - | 1e-118 | 49.38% (405) | amidohydrolase 2 |
| M. ulcerans Agy99 | MUL_3143 | - | 2e-86 | 44.92% (374) | amidohydrolase |
| M. vanbaalenii PYR-1 | Mvan_4211 | - | 0.0 | 79.58% (431) | amidohydrolase 2 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2441|M.gilvum_PYR-GCK ----------------MDKNDMILISVDDHIVEPPDMFKNHLSKKYLDEA
Mvan_4211|M.vanbaalenii_PYR-1 MSRAPARKGGNGRSAVMNRDDMILISVDDHIVEPPDMFKNHLPRKYLDEA
MMAR_4742|M.marinum_M ----------------MNKEDMILISVDDHTVEPPNMFKNHLSAKYLDDA
MAV_0907|M.avium_104 ----------------MNKEDMILISVDDHTVEPPDMFKNHLPKKYLDDA
MSMEG_4837|M.smegmatis_MC2_155 ----------------MNKDDMILISVDDHIIEPPDMFKNHLPAKYRDEA
TH_2339|M.thermoresistible__bu ------------MSHDLRPEDLILVSIDDHVVEPPDMFLRHVPAKYRDEA
MUL_3143|M.ulcerans_Agy99 -----------MTDPPRARRRYTVISVDDHIVEPPDTFTGRLPQRFADRA
::*:*** :***: * ::. :: * *
Mflv_2441|M.gilvum_PYR-GCK PRLVHNEDGSDTWQFRDVVIPNVALNAVAGRPKEEYGLEPQGLDEIRPGC
Mvan_4211|M.vanbaalenii_PYR-1 PRLVHNPDGSDTWQFRDVVIPNVALNAVAGRPKEEYGLEPQGLDEIRPGC
MMAR_4742|M.marinum_M PRLVHNPDGSDMWKFRDTVIPNVALNAVAGRPKEEYGLEPQGLDEIRPGC
MAV_0907|M.avium_104 PRLVHNPDGSDMWKFRDTVIPNVALNAVAGRPKEEYGIEPTGLDEIRPGC
MSMEG_4837|M.smegmatis_MC2_155 PRLVHNPDGSDTWQFRDTVIPNVALNAVAGRPKEEYGLEPQGLDEIRKGC
TH_2339|M.thermoresistible__bu PIVVTDENGVDQWMYQGRPQGVSGLNAVVSWPPEEWGRNPARFAEMRPGV
MUL_3143|M.ulcerans_Agy99 PRVVETDDGGQVWVYDGQVLPNVGFNAVVGRPVSEYGFEPVRFDEMRRGA
* :* :* : * : . .:***.. * .*:* :* : *:* *
Mflv_2441|M.gilvum_PYR-GCK WQVDERVKDMNAGGILGSMCFPSFP-GFAGR--LFATEDPEFSLALVQAY
Mvan_4211|M.vanbaalenii_PYR-1 WQVDERVKDMNAGGILGSMCFPSFP-GFAGR--LFATEDAEFSLALVQAY
MMAR_4742|M.marinum_M YNVDERIKDMNAGGILASICFPSFP-GFAGR--LFATEDPDFSVALVQAY
MAV_0907|M.avium_104 YNVDERVKDMNAGGILASICFPSFP-GFAGR--LFATEDNDFSIALVQAY
MSMEG_4837|M.smegmatis_MC2_155 YDPNERVKDMNAGGVLATMNFPSFP-GFAAR--LFATDDEEFSLALVQAY
TH_2339|M.thermoresistible__bu YDVHERVRDMNRNGILASMCFPTFT-GFSAR--HLNMHREHVTLIMVSAY
MUL_3143|M.ulcerans_Agy99 WDIHERIKDMDLNGVYASLNFPSFLPGFAGQRLQQVTKDPELAWAAVRAW
:: .**::**: .*: .:: **:* **:.: . ..: * *:
Mflv_2441|M.gilvum_PYR-GCK NDWHVEEWCGAYPARFIPMTLPVIWDPEACAKEIRRNAARGVHSLTFSEN
Mvan_4211|M.vanbaalenii_PYR-1 NDWHVEEWCGAYPARFIPMTLPVIWDPEACAKEIRRNAARGVHSLTFSEN
MMAR_4742|M.marinum_M NDWHIDEWCGAYPARFVPMAIPVIWDAEACAAEVRRVAKKGVHALTFTEN
MAV_0907|M.avium_104 NDWHIDEWCGAYPARFIPMAIPVIWDAEECAKEVRRVSKKGVHALTFTEN
MSMEG_4837|M.smegmatis_MC2_155 NDWHIDEWCASNPGRFIPMAIPAIWNPGLAAAEVRRVAEKGVHSLTFTEN
TH_2339|M.thermoresistible__bu NDWHIDEWAGSYPGRFIPIAILPTWNPEAMCAEIRRVAAKGCRAVTMPEL
MUL_3143|M.ulcerans_Agy99 NDWHLEAWAGPYPERIIPCQLPWLLDPSAGAQLIHENAERGFRAVSFSEN
****:: *... * *::* : :. . ::. : :* :::::.*
Mflv_2441|M.gilvum_PYR-GCK PSAMGYPSFHDFDHWKPMWDALVDTDTVLNVHIGSSGRLAITAPDAPMDV
Mvan_4211|M.vanbaalenii_PYR-1 PSAMGYPSFHDFEHWKPMWDALVDTDTVLNVHIGSSGRLAITAPDAPMDV
MMAR_4742|M.marinum_M PAAMGYPSFHN-EYWNPLWKALCDTNTVMNIHIGSSGRLAITAPDAPMDV
MAV_0907|M.avium_104 PAAMGYPSFHD-EYWNPLWKALVDTNTVMNVHIGSSGRLAITAPDAPMDV
MSMEG_4837|M.smegmatis_MC2_155 PSTLGYPSFHDLEYWRPLWQALVDTETVMNVHIGSSGKLAITAPDAPMDV
TH_2339|M.thermoresistible__bu PHLEGLPSYHDEDYWGPVFRTLSEENVVMCLHIGTGFGAISMAPDAPIDN
MUL_3143|M.ulcerans_Agy99 PAMLGLPSIHS-GHWDPMMAACAETGTVVNLHIGSSGSSPSTTDDAPADV
* * ** *. :* *: : : .*: :***:. : *** *
Mflv_2441|M.gilvum_PYR-GCK MITLQPMNIVQAAADLLWSRPIKEYPDLKIALSEGGTGWIPYFLERVDRT
Mvan_4211|M.vanbaalenii_PYR-1 MITLQPMNIVQAAADLLWSRPIKEYPDLKIALSEGGTGWIPYFLERVDRT
MMAR_4742|M.marinum_M MITLQPMNIVQAAADLLWSRPIKEYPDLKIALSEGGTGWIPYFLERADRT
MAV_0907|M.avium_104 MITLQPMNIVQAAADLLWSRPIKEFPDLKIALSEGGTGWIPYFLERADRT
MSMEG_4837|M.smegmatis_MC2_155 MITLQPMNIVQAAADLLWSAPIKEFPTLKIALSEGGTGWIPYFLDRVDRT
TH_2339|M.thermoresistible__bu MIILATQVSAMCAQDLLWGPAMRNYPDLKFAFSEGGIGWIPFYLDRCDRH
MUL_3143|M.ulcerans_Agy99 PGVLFFAYAISAAVDWLYSGLPSRFPDLKICLSEGRIGWVAGLLDRLDHM
* .* * *:. .:* **:.:*** **:. *:* *:
Mflv_2441|M.gilvum_PYR-GCK YEMHSTWTG-QDFKGKLP-SEVFREHFLTCFIADPVGVATRHQIGVDNIC
Mvan_4211|M.vanbaalenii_PYR-1 YEMHSTWTG-QDFKGKLP-SEVFREHFLTCFIADPVGVATRHQIGVDNIC
MMAR_4742|M.marinum_M FEMHSTWTH-QNFGGKIP-SEVFREHFLTCFISDKVGVALRNMIGIDNIA
MAV_0907|M.avium_104 FEMHSAWTH-QDFKGKLP-SEVFREHFLTCFISDKVGVALRNEIGIDNIC
MSMEG_4837|M.smegmatis_MC2_155 FEMHSTWTH-QNFGGKLP-SEVFREHFMTCFISDPVGVKNRHMIGIDNIC
TH_2339|M.thermoresistible__bu YTN-QKWLR-RDFGDKLP-SDVFREHSLACYVTDKTSLKLRHEIGIDIIA
MUL_3143|M.ulcerans_Agy99 LSYHEMYGTWRAMGESLTSAEVFTRNFWFCAVEDKSSFVQHYRIGTENIM
. : : : .:. ::** .: * : * .. : ** : *
Mflv_2441|M.gilvum_PYR-GCK WEADYPHSDSMWPGAPEQLDEVLKANNVPDDEINKMTYENAMRWYHWDPF
Mvan_4211|M.vanbaalenii_PYR-1 WEADYPHSDSMWPGAPEQLDEVLKVNSVPDDEIDKMTYQNAMRWYHWDPF
MMAR_4742|M.marinum_M WEADYPHSDSMWPGAPEELWDVLSVNDVPDGEINKMTYENAMRWYSFDPF
MAV_0907|M.avium_104 WEADYPHSDSMWPGAPEELWDVLSINNVPDDEINKMTYQNAMRWYSFDPF
MSMEG_4837|M.smegmatis_MC2_155 WEMDYPHSDSMWPGAPEELSAVFDTYDVPDDEINKITHENAMRLYHFDPF
TH_2339|M.thermoresistible__bu WECDYPHSDCFWPDAPEQVHAELRAAGADDFDINKITWENSCRFFSWDPF
MUL_3143|M.ulcerans_Agy99 VESDYPHCDSTWPHTQQTMHEQLDG--LPADVIRKITWENAARLYQHPVP
* ****.*. ** : : : : * *:* :*: * :
Mflv_2441|M.gilvum_PYR-GCK THIAKEQATVGALRKAAEGHDVSIQALSNKE--KTGTTFSDFAASAKSIS
Mvan_4211|M.vanbaalenii_PYR-1 THIPKEQATVGALRKAAEGHDVSIQALSHKE--KTGATFADFAARAKEIT
MMAR_4742|M.marinum_M SHITREQATVGALRKAAEGHDVSVRALSHHKDARSGSSVAEFTANAKALT
MAV_0907|M.avium_104 THISREQATVGALRKAAEGHDVSIKAQGHDKDSRGGSSFAEFAANAKALT
MSMEG_4837|M.smegmatis_MC2_155 AHVPKEQATVGALRKAAEGHDVSIRALSKKD--KTGASFADFQANAKAVS
TH_2339|M.thermoresistible__bu EHTPRAEATVGALRAKATDVDTTIRSR---------AEWAQLYEQKQQFA
MUL_3143|M.ulcerans_Agy99 VAIAQNPEAF----------------------------------------
.: :.
Mflv_2441|M.gilvum_PYR-GCK GNKD
Mvan_4211|M.vanbaalenii_PYR-1 GNTD
MMAR_4742|M.marinum_M GNKD
MAV_0907|M.avium_104 GNKD
MSMEG_4837|M.smegmatis_MC2_155 GARD
TH_2339|M.thermoresistible__bu QA--
MUL_3143|M.ulcerans_Agy99 ----