For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LRDVDREHIDMMVLYPSLGFCILRLDDPDFATRLARFYNQWIGDYCAPTNGWLRGGGVTSMERGQVAIDI TNGVKELGIAVTLIPPVLNASNLDHPYLGPFYAATVERGMAISIHARYPFAADWC*
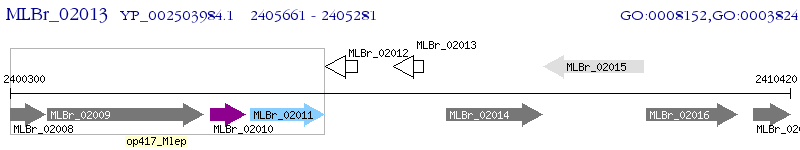
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_02013 | - | - | 100% (126) | hypothetical protein MLBr_02013 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2798 | - | 3e-39 | 61.21% (116) | amidohydrolase |
| M. avium 104 | MAV_2798 | - | 4e-37 | 58.62% (116) | amidohydrolase family protein |
| M. smegmatis MC2 155 | MSMEG_6487 | - | 2e-06 | 28.03% (132) | 2-hydroxy-3-carboxy-6-oxo-7-methylocta-2, 4-dienoate |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2798|M.marinum_M MRIIDADGHVAENASLTGEALRRWPDRVRLTTRGRPQLMIEGRHYPQDNG
MAV_2798|M.avium_104 MRIIDADGHVAENSSLAIEAIQRWPEHVKPGRHGRLGLMIEGRNYPEDGG
MLBr_02013|M.leprae_Br4923 --------------------------------------------------
MSMEG_6487|M.smegmatis_MC2_155 ----------MITAPLAGGVIDVHAHWLPRELFDLPPGAPYGPLHDRDGQ
MMAR_2798|M.marinum_M PGAGCPPQHGISKAPGINCATADGVLADADRDHLDTMVLYPSVGLCVPSL
MAV_2798|M.avium_104 PGAGCPPEHGISDAPDINCRSAEGVLGDADRDHIDTMVLYPSLGLCAPSL
MLBr_02013|M.leprae_Br4923 ------------------------MLRDVDREHIDMMVLYPSLGFCILRL
MSMEG_6487|M.smegmatis_MC2_155 LHLGDLPLS----IATDAMSDEAAIIDDMDRAGVGVRVLSAPP--FAFPL
:: * ** :. ** .. *
MMAR_2798|M.marinum_M EDPAFAAGFARLYNEWIADFCAPTNGRLRGVGVTPIEH------GQVAID
MAV_2798|M.avium_104 RDPEFAAGFARLYNRWIADYCAQSGGRLRGVAVTPIEH------GQLAID
MLBr_02013|M.leprae_Br4923 DDPDFATRLARFYNQWIGDYCAPTNGWLRGGGVTSMER------GQVAID
MSMEG_6487|M.smegmatis_MC2_155 RGGDDADRYAAEFNQALAKLVARGDGRLAGLGIVSLDAGPDFASGSVPRQ
. * * :*. :.. * .* * * .:..:: *.:. :
MMAR_2798|M.marinum_M IMKEANELGLVATLVPPALKSRNLDHADLDPFYAAAVEQGMPLGIHGAPG
MAV_2798|M.avium_104 VMREAKDLGLVATLVPPALQSRNLDHPDLDPFYAAAVELGMPLGVHGAPG
MLBr_02013|M.leprae_Br4923 ITNGVKELGIAVTLIPPVLNASNLDHPYLGPFYAATVERGMAISIHAR--
MSMEG_6487|M.smegmatis_MC2_155 LDHLAAVDGIAGVAIPPVVDGRSFDGAALRHVLTETARRDLAVLVHPMQ-
: . . *:. . :**.:.. .:* . * . : :.. .:.: :*
MMAR_2798|M.marinum_M MHLPKIGVDRFTNYIQVHCISFPFDQMTAMTALVSGGVFDRHPRLRVAFL
MAV_2798|M.avium_104 IHLPKIGVDRFTNYIQVHCISFPFDQMTAMTAMVSGGVFERHPKLRVAFL
MLBr_02013|M.leprae_Br4923 ---------------------YPF--------------------------
MSMEG_6487|M.smegmatis_MC2_155 -----IGRAEWSQFYLANLIGNPVETATAVATLILSGLQEELPDLRICFL
*.
MMAR_2798|M.marinum_M EAGVGWVPFFVDRLHEHYEKRGDWIDGGWRRDPHDYLRAGNVWVTCEPDE
MAV_2798|M.avium_104 EAGAGWVPFFIDRLHEHYEKRGDWVERGWRRDPHEYLKAGNIFVTCEPEE
MLBr_02013|M.leprae_Br4923 --AADWC-------------------------------------------
MSMEG_6487|M.smegmatis_MC2_155 H-GGGCAPGLLGRWSHGWTARADVRSRTSRPPAEAFRRLY--FDTITHGV
. .
MMAR_2798|M.marinum_M PILPGVIDVLGDDFIMFASDYPHWDGEWPESTKQLRTRTDIGEQSRNKIA
MAV_2798|M.avium_104 PILPGVIDVLGADFIMFASDYPHWDGEWPESTKHLRTRADISEETREKIG
MLBr_02013|M.leprae_Br4923 --------------------------------------------------
MSMEG_6487|M.smegmatis_MC2_155 PQLELLTELAGEDRIVCGSDYPFDMAEVDPARFAVEHGPGQDSLIRAARA
MMAR_2798|M.marinum_M GLNAQRFYELN-------------------
MAV_2798|M.avium_104 GLNAQRFYNLN-------------------
MLBr_02013|M.leprae_Br4923 ------------------------------
MSMEG_6487|M.smegmatis_MC2_155 FLGIREARAGDTINPYRNPSATPVLAPDAT