For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RIIDADGHVAENSSLAIEAIQRWPEHVKPGRHGRLGLMIEGRNYPEDGGPGAGCPPEHGISDAPDINCRS AEGVLGDADRDHIDTMVLYPSLGLCAPSLRDPEFAAGFARLYNRWIADYCAQSGGRLRGVAVTPIEHGQL AIDVMREAKDLGLVATLVPPALQSRNLDHPDLDPFYAAAVELGMPLGVHGAPGIHLPKIGVDRFTNYIQV HCISFPFDQMTAMTAMVSGGVFERHPKLRVAFLEAGAGWVPFFIDRLHEHYEKRGDWVERGWRRDPHEYL KAGNIFVTCEPEEPILPGVIDVLGADFIMFASDYPHWDGEWPESTKHLRTRADISEETREKIGGLNAQRF YNLN*
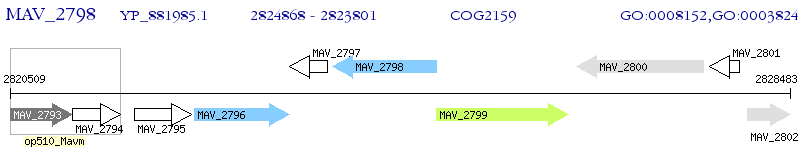
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2798 | - | - | 100% (355) | amidohydrolase family protein |
| M. avium 104 | MAV_1917 | - | 4e-25 | 29.02% (286) | amidohydrolase 2 |
| M. avium 104 | MAV_2629 | - | 2e-24 | 29.02% (286) | amidohydrolase 2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4324 | - | 2e-15 | 23.66% (262) | amidohydrolase 2 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_02013 | - | 8e-37 | 58.62% (116) | hypothetical protein MLBr_02013 |
| M. abscessus ATCC 19977 | MAB_0299 | - | 5e-11 | 23.14% (229) | putative amidohydrolase (aminocarboxymuconate-semialdehyde |
| M. marinum M | MMAR_2798 | - | 0.0 | 81.13% (355) | amidohydrolase |
| M. smegmatis MC2 155 | MSMEG_4837 | - | 1e-18 | 26.69% (266) | amidohydrolase 2 |
| M. thermoresistible (build 8) | TH_2827 | - | 3e-19 | 27.79% (349) | PUTATIVE amidohydrolase 2 |
| M. ulcerans Agy99 | MUL_0389 | - | 1e-17 | 26.32% (304) | hypothetical protein MUL_0389 |
| M. vanbaalenii PYR-1 | Mvan_0901 | - | 4e-20 | 25.68% (292) | amidohydrolase 2 |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_2798|M.avium_104 -----MRIIDADGHVAENSSLAIEAIQR----WPEHVKPG------RHGR
MMAR_2798|M.marinum_M -----MRIIDADGHVAENASLTGEALRR----WPDRVRLT------TRGR
MLBr_02013|M.leprae_Br4923 --------------------------------------------------
Mflv_4324|M.gilvum_PYR-GCK MNRDDMILISVDDHIVEPPDMFKNHLSKKYLVEAPRLVHN------EDGS
MSMEG_4837|M.smegmatis_MC2_155 MNKDDMILISVDDHIIEPPDMFKNHLPAKYRDEAPRLVHN------PDGS
TH_2827|M.thermoresistible__bu VTAPALEIWDADNHLYEPTDAYTRHLPKQYRGAIQWIQVDGLTKLMIKGR
Mvan_0901|M.vanbaalenii_PYR-1 -MGSSTLIIDTDTHITEPPDLWTSRMSKSRWGNLIPEVKWVE----EKQG
MUL_0389|M.ulcerans_Agy99 --MDRYTVISADCHAGADLFDYREYLEAKY--------------------
MAB_0299|M.abscessus_ATCC_1997 -----MTSVRIDIHAHLWSDEYLSLLDG----------------------
MAV_2798|M.avium_104 LGLMIEGRNYPEDGGPGAGCPPEHGISDAPD------------INCRSAE
MMAR_2798|M.marinum_M PQLMIEGRHYPQDNGPGAGCPPQHGISKAPG------------INCATAD
MLBr_02013|M.leprae_Br4923 --------------------------------------------------
Mflv_4324|M.gilvum_PYR-GCK DTWQFRDVVIPNVALNAVAGRPKEEYGLEPQGLDEIR------PGCWQVD
MSMEG_4837|M.smegmatis_MC2_155 DTWQFRDTVIPNVALNAVAGRPKEEYGLEPQGLDEIR------KGCYDPN
TH_2827|M.thermoresistible__bu LTDTIPNPTYEVVASPGAWADYFRGINPEGKSLRELANPIRCPEEFRHAE
Mvan_0901|M.vanbaalenii_PYR-1 EYWCMDGQPVFTVGTCIMVPGEDGKPVRSPRFPDYADRFSSMHPSAYDAK
MUL_0389|M.ulcerans_Agy99 -QDEFDDWAKTFVNPFGDLSEPDAEHN-------------------WDSD
MAB_0299|M.abscessus_ATCC_1997 ----YGRPATDVHRGLGAGATRD------------------------DLD
MAV_2798|M.avium_104 GVLGDADRDHIDTMVLYPSLG-----------LCAPSLRDPEFAAGFARL
MMAR_2798|M.marinum_M GVLADADRDHLDTMVLYPSVG-----------LCVPSLEDPAFAAGFARL
MLBr_02013|M.leprae_Br4923 -MLRDVDREHIDMMVLYPSLG-----------FCILRLDDPDFATRLARF
Mflv_4324|M.gilvum_PYR-GCK ERVKDMNAGGILGSMCFPSFPGF--------AGRLFATEDPEFSLALVQA
MSMEG_4837|M.smegmatis_MC2_155 ERVKDMNAGGVLATMNFPSFPGF--------AARLFATDDEEFSLALVQA
TH_2827|M.thermoresistible__bu QRLRLLDAQGIQACVMFPTTGG---------MLEERMLDDIELTHAVVHA
Mvan_0901|M.vanbaalenii_PYR-1 ARLEVMDAYGIQAAAIFPNLGFVGP-----NIFAAAGPDALEFQTAALQA
MUL_0389|M.ulcerans_Agy99 RRNAELDSDGVAGEVIFPNTVPPFFPQGSLAAPAPSTARDLELRWAGLRA
MAB_0299|M.abscessus_ATCC_1997 GRFELMDEAGVDLQILTATPAS-------------PHFEDQAAAVASARF
: : .. :
MAV_2798|M.avium_104 YNRW-IADYCAQSGGRLRGVAVTPIEHGQLAIDVMR-EAKDLGLVATLVP
MMAR_2798|M.marinum_M YNEW-IADFCAPTNGRLRGVGVTPIEHGQVAIDIMK-EANELGLVATLVP
MLBr_02013|M.leprae_Br4923 YNQW-IGDYCAPTNGWLRGGGVTSMERGQVAIDITN-GVKELGIAVTLIP
Mflv_4324|M.gilvum_PYR-GCK YNDWHVEEWCGAYPARFIPMTLPVIWDPEACAKEIR-RNAARGVHSLTFS
MSMEG_4837|M.smegmatis_MC2_155 YNDWHIDEWCASNPGRFIPMAIPAIWNPGLAAAEVR-RVAEKGVHSLTFT
TH_2827|M.thermoresistible__bu YNEWLLEEWTFDYAGRIFATPVITLPDVDKAVAELN-WCVENGARAVLVN
Mvan_0901|M.vanbaalenii_PYR-1 YNDF-LLDWSSIAPDRLLSLALIPYWDVDAAVAEIE-RCAAAGHKGLVST
MUL_0389|M.ulcerans_Agy99 HNRW-LADFCSLSPERRAGVGQILLGDVDEAVAEVA-QIAKLGLPGGVLL
MAB_0299|M.abscessus_ATCC_1997 INDE-YAKLSSEYPGRFGALASLPLPHVPAALEELARAIDELGMYGASIT
* . . *
MAV_2798|M.avium_104 PALQSR------NLDHPDLDPFYAAAVELGMPLGVHGAPGIHL-------
MMAR_2798|M.marinum_M PALKSR------NLDHADLDPFYAAAVEQGMPLGIHGAPGMHL-------
MLBr_02013|M.leprae_Br4923 PVLNAS------NLDHPYLGPFYAATVERGMAISIHAR------------
Mflv_4324|M.gilvum_PYR-GCK ENPSAMGY--PSFHDFDHWKPMWDALVDTDTVLNVHIGSS----------
MSMEG_4837|M.smegmatis_MC2_155 ENPSTLGY--PSFHDLEYWRPLWQALVDTETVMNVHIGSS----------
TH_2827|M.thermoresistible__bu PRPVATITGHTTSMGQPYFDPFWHRVEQLGIPVLMHACDSGYDRYSRDWE
Mvan_0901|M.vanbaalenii_PYR-1 GKPHEHGY---KLLADRYWDPMWAAAQDANLSISFHVGGGNLSRHLND--
MUL_0389|M.ulcerans_Agy99 PGVAPGTG--IPALYAEHWEPLWAACAQAGLVINHHGGNAG---------
MAB_0299|M.abscessus_ATCC_1997 TSVLGR------SIADPIFDPLYAELNRRRAVLFVHPAGCG---------
*:: : *
MAV_2798|M.avium_104 --PKIGVDRFTNYIQVHCISFPFDQMTAMTAMVSGGVFERHPKLRVAFLE
MMAR_2798|M.marinum_M --PKIGVDRFTNYIQVHCISFPFDQMTAMTALVSGGVFDRHPRLRVAFLE
MLBr_02013|M.leprae_Br4923 --------------------YPF---------------------------
Mflv_4324|M.gilvum_PYR-GCK --GRLAITAPDAPMDVMITLQPMNIVQAAADLLWSRPIKEYPDLKIALSE
MSMEG_4837|M.smegmatis_MC2_155 --GKLAITAPDAPMDVMITLQPMNIVQAAADLLWSAPIKEFPTLKIALSE
TH_2827|M.thermoresistible__bu GKGGEYTPFKPDAFRTIVYEDARSILDTCAALVAHGVFTRHPGVRVGVVE
Mvan_0901|M.vanbaalenii_PYR-1 --ERTSVEGWRATLARLTTSFFLESGITLADLLMSGVLARYPKLRFVSVE
MUL_0389|M.ulcerans_Agy99 ---PSPIDGWGSSLAVLVYESHWWAHRALLHLIFSGVLDRYPDLTVVLTE
MAB_0299|M.abscessus_ATCC_1997 ----AESSLITDHSLTWSIGAPIEDTVAIMHLIVAGVPSRYPDMKIVTCH
MAV_2798|M.avium_104 AGAGWVPFFIDRLHEHYEKRGDWVER---------GWRRDPHEYLKAGNI
MMAR_2798|M.marinum_M AGVGWVPFFVDRLHEHYEKRGDWIDG---------GWRRDPHDYLRAGNV
MLBr_02013|M.leprae_Br4923 -------------------AADWC--------------------------
Mflv_4324|M.gilvum_PYR-GCK GGTGWIPYFLERVDRTYEMHSTWTGQ---------DFKGKLPSEVFR-EH
MSMEG_4837|M.smegmatis_MC2_155 GGTGWIPYFLDRVDRTFEMHSTWTHQ---------NFGGKLPSEVFR-EH
TH_2827|M.thermoresistible__bu NGGAWVPRLIDLFERVYKKMPNEFG--------------AHPVEQFREHV
Mvan_0901|M.vanbaalenii_PYR-1 SAVGWIPFLLESLDFHYKKYEPWLERPE------FAGNDMLPSDYFHRQV
MUL_0389|M.ulcerans_Agy99 QGTGWIPATLDSLDVAAARYRRPNSAIARFAGPTAGSLPLQPSQYWARQC
MAB_0299|M.abscessus_ATCC_1997 LG-GALPMVLERAHRQVTEWEATQCP------------EAPRDAARRLWY
MAV_2798|M.avium_104 FVTCEPEEPILPGVIDVLGADFIMFASDYPHWDGEWP---ESTKHLRTRA
MMAR_2798|M.marinum_M WVTCEPDEPILPGVIDVLGDDFIMFASDYPHWDGEWP---ESTKQLRTRT
MLBr_02013|M.leprae_Br4923 --------------------------------------------------
Mflv_4324|M.gilvum_PYR-GCK FLTCFIADPVGVATRHQIGVDNICWEADYPHSDSMWPGAPEQLDEVLKAN
MSMEG_4837|M.smegmatis_MC2_155 FMTCFISDPVGVKNRHMIGIDNICWEMDYPHSDSMWPGAPEELSAVFDTY
TH_2827|M.thermoresistible__bu WVNPFHEEDLS-GLIDLIGADRVLFGSDFPHPEGLAEP----AKLTPEIA
Mvan_0901|M.vanbaalenii_PYR-1 YAN-YWYEDLQPWHVAAIGEDNLMFETDYPHQTCLDKEE-IQHSINEGLS
MUL_0389|M.ulcerans_Agy99 YVGASFLRPVECAQRHRIGVDRIMWASDYPHMEGTAPYS--REALRHTFS
MAB_0299|M.abscessus_ATCC_1997 DTVGHDHTPALRAAVASLGVDRLIFGSDFPYQAGPRYVS----SATYIEE
MAV_2798|M.avium_104 DISEETREKIGGLNAQRFYNLN----------------------------
MMAR_2798|M.marinum_M DIGEQSRNKIAGLNAQRFYELN----------------------------
MLBr_02013|M.leprae_Br4923 --------------------------------------------------
Mflv_4324|M.gilvum_PYR-GCK NVPDDEINKMTYENAMRWYHWDPFTHIAKEQATVGALRKAAEGHDVSIQA
MSMEG_4837|M.smegmatis_MC2_155 DVPDDEINKITHENAMRLYHFDPFAHVPKEQATVGALRKAAEGHDVSIRA
TH_2827|M.thermoresistible__bu ELPEPTIEAVMGGNLRELLTAVPHGR------------------------
Mvan_0901|M.vanbaalenii_PYR-1 GVSETTKEKILWRNAASLFNMDIAKLEKLT--------------------
MUL_0389|M.ulcerans_Agy99 DVDPDEVAAMVGGNAAAVYGFDLQALAPLAARIGPTVTEVAEPLAAIPAD
MAB_0299|M.abscessus_ATCC_1997 GLSAEDASAILDHNGAELLGLTGV--------------------------
MAV_2798|M.avium_104 -----------------------------
MMAR_2798|M.marinum_M -----------------------------
MLBr_02013|M.leprae_Br4923 -----------------------------
Mflv_4324|M.gilvum_PYR-GCK LSHHKEGERGGGAGHFDALNAAARGNSGA
MSMEG_4837|M.smegmatis_MC2_155 LS--KKDKTGASFADFQANAKAVSGARD-
TH_2827|M.thermoresistible__bu -----------------------------
Mvan_0901|M.vanbaalenii_PYR-1 -----------------------------
MUL_0389|M.ulcerans_Agy99 ASSTAFEPDPIRAW---------------
MAB_0299|M.abscessus_ATCC_1997 -----------------------------