For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSTPTVPSPAHPPTLYPPEGFGAPKHRHGHSTGAVTGLPADTVIFSADNHISVADDIFYERFPEELKGAA PRIWYEDGAYMVGMKGKAWTGGDFGRVLMQYDDLAGAASNNIEARIRELKEDGIDKELAFPNAVLALFHY PDKALRERVFRIYNEHIADLQERSNGHFYGVGLINWWDPKGTRSTLEELKSLGLKTFLLPLNPGKDDEGN IYDYGSTSMDAVWDEIEAAGLPVSHHIGETPPKTPCQNNSVVVGMMVNVDSFREQFAKYVFSGILDRHPS LKIGWFEGGIAWVPTALQDAEHMLASYRHMFNHELQHDVNHYWANHMSASFMVDPLGLQLIDRIGVDNVM WSSDYPHNESTFGYSEKSLATVVDAVGPEAAVKIVSSNVQRFLGL
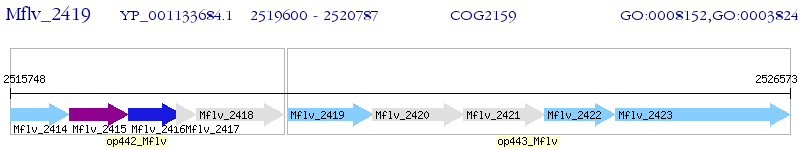
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2419 | - | - | 100% (395) | amidohydrolase 2 |
| M. gilvum PYR-GCK | Mflv_2441 | - | 7e-18 | 21.15% (383) | amidohydrolase 2 |
| M. gilvum PYR-GCK | Mflv_2371 | - | 7e-18 | 22.19% (383) | amidohydrolase 2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4761 | - | 0.0 | 75.00% (388) | hypothetical protein MMAR_4761 |
| M. avium 104 | MAV_0890 | - | 1e-170 | 77.62% (353) | amidohydrolase family protein |
| M. smegmatis MC2 155 | MSMEG_4855 | - | 0.0 | 90.08% (383) | amidohydrolase family protein |
| M. thermoresistible (build 8) | TH_2277 | - | 0.0 | 84.63% (397) | amidohydrolase family protein |
| M. ulcerans Agy99 | MUL_0389 | - | 3e-23 | 28.34% (307) | hypothetical protein MUL_0389 |
| M. vanbaalenii PYR-1 | Mvan_4233 | - | 0.0 | 94.26% (383) | amidohydrolase 2 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4761|M.marinum_M MSAPTSS-----VSLYPPEGFGAPKNRRGHA--SGINVGLPEGTVVFSAD
MAV_0890|M.avium_104 --------------------------------------------MVFSAD
Mflv_2419|M.gilvum_PYR-GCK MSTPTVPSPAHPPTLYPPEGFGAPKHRHGHS-TGAVTG-LPADTVIFSAD
Mvan_4233|M.vanbaalenii_PYR-1 MSIRTTAATAT-ATLYPPEGFGAPKNRHGHAAEGGLTG-LPEGTEIFSAD
MSMEG_4855|M.smegmatis_MC2_155 MSVAT-------QTLYPPEGFGAPKHRRGHASPDGLAS-LPSGTEIFSAD
TH_2277|M.thermoresistible__bu MSTPT--------LVYPAEGYGAPKNRRGRAPRDVTELGLPAGTEVFSAD
MUL_0389|M.ulcerans_Agy99 ----------------------------------------MDRYTVISAD
::***
MMAR_4761|M.marinum_M NHISLAADIFYERFPDDLKDKAPRIWYEDGAFQVGRKGQSFLPGDFSAVL
MAV_0890|M.avium_104 NHISLADDIFYQRFPDDLKDKAPRIWYEDGAYQVGRKGQSFLPGDFSAVL
Mflv_2419|M.gilvum_PYR-GCK NHISVADDIFYERFPEELKGAAPRIWYEDGAYMVGMKGKAWTGGDFGRVL
Mvan_4233|M.vanbaalenii_PYR-1 NHISVADDIFYERFPEELKGAAPRIWYEDGAYMVGMKGKAWTGGDFGRVL
MSMEG_4855|M.smegmatis_MC2_155 NHISVADDIFYDRFPEELKGAAPRIWYEDGAYMVGMKGKAWTGGDFGRVL
TH_2277|M.thermoresistible__bu NHISVAEDIFYERFPDDLKGAAPRIWYEDGAYMVGMKGKSWVGGDFGRVL
MUL_0389|M.ulcerans_Agy99 CHAG-ADLFDYREYLEAKYQDEFDDWAKT----------------FVNPF
* . * : * .: : * : * :
MMAR_4761|M.marinum_M MQYDDLPGAASTNIEARIQELREDGVDKELAFPNAVLALFHYP-------
MAV_0890|M.avium_104 MQYDDLPGAASTNIEARIQELHEDGVEKELAFPNAVLALFHYP-------
Mflv_2419|M.gilvum_PYR-GCK MQYDDLAGAASNNIEARIRELKEDGIDKELAFPNAVLALFHYP-------
Mvan_4233|M.vanbaalenii_PYR-1 MQYDDLAGAASNNIEARIRELKEDGIDKELAFPNAVLALFHYP-------
MSMEG_4855|M.smegmatis_MC2_155 MQYDDLAGAASNNIAARVRELKEDGIDKELAFPNAVLALFHYP-------
TH_2277|M.thermoresistible__bu MQYDDLAGAATNNIEARVQELAEDGITRELAFPNAVLALFHYP-------
MUL_0389|M.ulcerans_Agy99 GDLSEPDAEHNWDSDRRNAELDSDGVAGEVIFPNTVPPFFPQGSLAAPAP
: .: . . : * ** .**: *: ***:* .:*
MMAR_4761|M.marinum_M ----DKMLRELAFRIYNEYIAELQERSAGHFYGAGLINWWDPDGARRTLE
MAV_0890|M.avium_104 ----DKTLRELAFRIYNEYIAELQERSNGHFYGAGLINWWDPQGTRKTLD
Mflv_2419|M.gilvum_PYR-GCK ----DKALRERVFRIYNEHIADLQERSNGHFYGVGLINWWDPKGTRSTLE
Mvan_4233|M.vanbaalenii_PYR-1 ----DKALRERVFRIYNEHIAELQERSNGHFYGVGLINWWDPKGTRSTLE
MSMEG_4855|M.smegmatis_MC2_155 ----DKGIRERVFRIYNEHIAALQEESKGHFYGVGLINWWDPSGTRSTLE
TH_2277|M.thermoresistible__bu ----DKALRERVFRIYNEHIADLQDRSNGHFYGVGLINWWDPKGTRSTLE
MUL_0389|M.ulcerans_Agy99 STARDLELRWAGLRAHNRWLADFCSLSPERRAGVGQILLGDVDEAVAEVA
* :* :* :*. :* : . * : *.* * * . : :
MMAR_4761|M.marinum_M ELKSLGLKTFLMPLNPGKDDDGNPIDYASTTMRPVWDEIEASGMPITHHI
MAV_0890|M.avium_104 ELKALGLKTFLMPLNPGKDDDGNMIDYSSTAMRPVWDEIEAAGLPVTHHI
Mflv_2419|M.gilvum_PYR-GCK ELKSLGLKTFLLPLNPGKDDEGNIYDYGSTSMDAVWDEIEAAGLPVSHHI
Mvan_4233|M.vanbaalenii_PYR-1 ELKSLGLRTFLLPLNPGKDDDGNIYDYGSTEMDAVWDEIEAAGLPVSHHI
MSMEG_4855|M.smegmatis_MC2_155 QLKSLGLRTFLLPLNPGKDDEGNIYDYGSTAMDAVWDEIEASGIPVSHHI
TH_2277|M.thermoresistible__bu QLKSLGLKTFLLPLNPGKDDDGNIYDYGSRAMDPVWDEIEEAGLPVTHHI
MUL_0389|M.ulcerans_Agy99 QIAKLGLPGGVL--LPGVAPGTGIPALYAEHWEPLWAACAQAGLVINHHG
:: *** :: ** . : .:* :*: :.**
MMAR_4761|M.marinum_M GETPPKSPCEFNSVVVGMMINIDGFR-ETFSKYIFGGILDQHPGLRVGWF
MAV_0890|M.avium_104 GETPPKSPTEFNSVVVGMMINIDGFR-ETFSKYIFGGILDDHPGLRIGWF
Mflv_2419|M.gilvum_PYR-GCK GETPPKTPCQNNSVVVGMMVNVDSFR-EQFAKYVFSGILDRHPSLKIGWF
Mvan_4233|M.vanbaalenii_PYR-1 GETPPKTPCQNNSVVVGMMVNVDSFR-EQFAKYVFSGILDRHPALKIGWF
MSMEG_4855|M.smegmatis_MC2_155 GETPPKTPCENNSVVVGMMVNVDSFR-EQFAKYVFSGILDRHPELKIGWF
TH_2277|M.thermoresistible__bu GETPPKTPCEHNSVVVGMMVNVDSFR-EQFSKYVFSGILDRHPGLRIGWF
MUL_0389|M.ulcerans_Agy99 GNAGPSPIDGWGSSLAVLVYESHWWAHRALLHLIFSGVLDRYPDLTVVLT
*:: *.. .* :. :: : . : . : : :*.*:** :* * :
MMAR_4761|M.marinum_M EGGIAWVPWALQDAEHLVASYQ------HMFN----RPLEHDVRYYWDKH
MAV_0890|M.avium_104 EGGISWVPWALQDAEHLVASYQ------HMFN----RPLQHDVRYYWDTH
Mflv_2419|M.gilvum_PYR-GCK EGGIAWVPTALQDAEHMLASYR------HMFN----HELQHDVNHYWANH
Mvan_4233|M.vanbaalenii_PYR-1 EGGIAWVPTALQDAEHMLASYR------HMFN----HELAHDVNHYWAHH
MSMEG_4855|M.smegmatis_MC2_155 EGGIAWVPTALQDAEHMLASYR------HMFN----HELQHDIRHYWDKH
TH_2277|M.thermoresistible__bu EGGIAWVPTALQDAEHLLASYR------HMFN----HELQHDIRHYWDNH
MUL_0389|M.ulcerans_Agy99 EQGTGWIPATLDSLDVAAARYRRPNSAIARFAGPTAGSLPLQPSQYWARQ
* * .*:* :*:. : * *: * * : ** :
MMAR_4761|M.marinum_M MSA-SFMVDPLGLELIDRIGVGKVMWSSDYPHNESTYGYSEKSLAAVVDA
MAV_0890|M.avium_104 MSA-SFMVDPLGLRLIDQIGVDKVMWSSDYPHNESTYGYSEKSLKAVVDA
Mflv_2419|M.gilvum_PYR-GCK MSA-SFMVDPLGLQLIDRIGVDNVMWSSDYPHNESTFGYSEKSLATVVDA
Mvan_4233|M.vanbaalenii_PYR-1 MSA-SFMVDPLGLQLIDRIGVDNVMWSSDYPHNESTFGYSEKSLASVVET
MSMEG_4855|M.smegmatis_MC2_155 MCA-SFMVDPLGLRLIDDIGVDNVMWSSDYPHNESTFGYSEKSLATVVDA
TH_2277|M.thermoresistible__bu MSA-SFMVDPLGLELIDRIGVDNVFWSSDYPHNESTYGYSEKSLAAVVQA
MUL_0389|M.ulcerans_Agy99 CYVGASFLRPVECAQRHRIGVDRIMWASDYPHMEGTAPYSREALRHTFSD
. : :: *: . ***..::*:***** *.* **.::* ...
MMAR_4761|M.marinum_M VGPQHAARIVSGNITEFLGL------------------------------
MAV_0890|M.avium_104 VGPANAAQIVSGNVTKFLGL------------------------------
Mflv_2419|M.gilvum_PYR-GCK VGPEAAVKIVSSNVQRFLGL------------------------------
Mvan_4233|M.vanbaalenii_PYR-1 VGAEAAVKIVSTNVQKFLGLRTQGAIGAEA--------------------
MSMEG_4855|M.smegmatis_MC2_155 VGPENAVKIVSTNIQKFLGLQ-----------------------------
TH_2277|M.thermoresistible__bu VGPENAVKIVSTNIQRFLGIAE----------------------------
MUL_0389|M.ulcerans_Agy99 VDPDEVAAMVGGNAAAVYGFDLQALAPLAARIGPTVTEVAEPLAAIPADA
*.. .. :*. * . *:
MMAR_4761|M.marinum_M -------------
MAV_0890|M.avium_104 -------------
Mflv_2419|M.gilvum_PYR-GCK -------------
Mvan_4233|M.vanbaalenii_PYR-1 -------------
MSMEG_4855|M.smegmatis_MC2_155 -------------
TH_2277|M.thermoresistible__bu -------------
MUL_0389|M.ulcerans_Agy99 SSTAFEPDPIRAW