For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRPDGPGFFAELMYAVNAAPWDIGGPQPVIRQLVALGAVRGEVLDPGCGTGWHAIEYARAGCSVTGVDLA PTAIARARNNTRTAGVEAQFVLGDATTLDYEARFDTVVDSKLLDNLDSAEARGRYLRSLYRAMKPQGRLF LYGFSDGHVNGFHNHELDGIDYEMALTAAGFTITYLGETTYRLSIPSYRRICTDCPDQVPPDGQIHIPMI EIHAVRNGLWQA
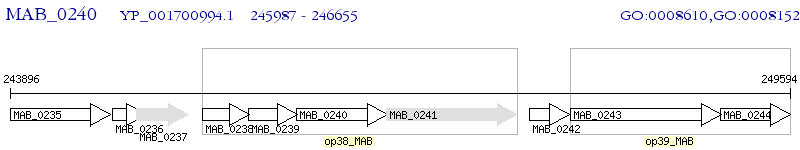
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0240 | - | - | 100% (222) | hypothetical protein MAB_0240 |
| M. abscessus ATCC 19977 | MAB_0867c | - | 9e-31 | 38.79% (214) | putative transferase |
| M. abscessus ATCC 19977 | MAB_0868c | - | 2e-29 | 40.00% (190) | hypothetical protein MAB_0868c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2694c | - | 2e-45 | 45.91% (220) | hypothetical protein Mb2694c |
| M. gilvum PYR-GCK | Mflv_4471 | - | 9e-24 | 40.49% (163) | methyltransferase type 11 |
| M. tuberculosis H37Rv | Rv2675c | - | 3e-45 | 45.91% (220) | hypothetical protein Rv2675c |
| M. leprae Br4923 | MLBr_01723 | - | 4e-07 | 30.10% (103) | hypothetical protein MLBr_01723 |
| M. marinum M | MMAR_2041 | - | 2e-42 | 46.36% (220) | methyltransferase |
| M. avium 104 | MAV_3570 | - | 3e-45 | 47.27% (220) | thiopurine S-methyltransferase (tpmt) superfamily protein |
| M. smegmatis MC2 155 | MSMEG_2067 | - | 6e-24 | 38.60% (171) | methyltransferase type 12 |
| M. thermoresistible (build 8) | TH_1722 | - | 2e-22 | 40.54% (148) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3309 | - | 3e-42 | 45.91% (220) | methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_1888 | - | 3e-27 | 41.86% (172) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2694c|M.bovis_AF2122/97 -------------------MTAQFDPADPTRFEEMYRDDRVAHGLPAATP
Rv2675c|M.tuberculosis_H37Rv -------------------MTAQFDPADPTRFEEMYRDDRVAHGLPAATP
MAV_3570|M.avium_104 -------------------MTASFDPADPARFEEMYRDQRTSHGLPAATP
MMAR_2041|M.marinum_M -------------------MTAP-DPTDPTRFEEMYRDQRVAHGLPAATP
MUL_3309|M.ulcerans_Agy99 -------------------MTAP-DPTDPTRFEEMYRDQRVAHGLPAATP
MAB_0240|M.abscessus_ATCC_1997 -----------------------MRPDGPGFFAELMYAVN-------AAP
Mflv_4471|M.gilvum_PYR-GCK --------------------MDMDQTPTLSRFDDFYKDED------KTPP
Mvan_1888|M.vanbaalenii_PYR-1 ----------------------MDLTPRLSRFDEFYKN--------QTPP
MSMEG_2067|M.smegmatis_MC2_155 ----------------------MDTTPTRELFDEAYES--------RTAP
TH_1722|M.thermoresistible__bu ---------------MATEDPGTGDPTDTIDWDSVYRQEAPG--FDGPPP
MLBr_01723|M.leprae_Br4923 MTRSTEISANAVPNPHATAEQVAAARKDSKLAQVLYHDWEAEN---YDEK
Mb2694c|M.bovis_AF2122/97 WDIGGPQ-----------PVVQQLVALGAIRGEVLDPGTGPGHHAIYYAA
Rv2675c|M.tuberculosis_H37Rv WDIGGPQ-----------PVVQQLVALGAIRGEVLDPGTGPGHHAIYYAA
MAV_3570|M.avium_104 WDIGGPQ-----------PVVRQLVALGAVKGEVLDPGTGPGHHAIYYAS
MMAR_2041|M.marinum_M WDIGGPQ-----------PVVEQLVALGAVKGEVLDPGTGPGHHAIYYAS
MUL_3309|M.ulcerans_Agy99 WDIGGPQ-----------PVVEQLVALGAVKGEVLDPGTGPGHHAIYYAS
MAB_0240|M.abscessus_ATCC_1997 WDIGGPQ-----------PVIRQLVALGAVRGEVLDPGCGTGWHAIEYAR
Mflv_4471|M.gilvum_PYR-GCK WVIGEPQ-----------PAVVDIERAGLITGRVLDVGCGTGERTILLTA
Mvan_1888|M.vanbaalenii_PYR-1 WVIGEPQ-----------QAIVELEQAGLIGGRVLDVGCGTGEHTILLAR
MSMEG_2067|M.smegmatis_MC2_155 WVIGEPQ-----------PAVVELERAGLIRSRVLDVGCGAGEHTILLTR
TH_1722|M.thermoresistible__bu WNIGEPQ-----------PELAALHRAGAFRSDVLDAGCGHAEISMALAA
MLBr_01723|M.leprae_Br4923 WSISYDQRCVDYVRHCFDAIVPDELFTELPYDCALELGCGTGFFLLNLIQ
* *. * : . .*: * * . :
Mb2694c|M.bovis_AF2122/97 KGYAATG--IDGSVAAIERARDNARKAGVSVN-FQVGDATTLDG-LDGRF
Rv2675c|M.tuberculosis_H37Rv KGYAATG--IDGSVAAIERARDNARKAGVSVN-FQVGDATTLDG-LDGRF
MAV_3570|M.avium_104 QGFSATG--IDGSAAAIERARANARTAGVSVN-FELADATKLDG-FDGRF
MMAR_2041|M.marinum_M KGYSATG--IDGSATAIERARDNAAKAGVSVN-FQVADATRLEG-LDNRF
MUL_3309|M.ulcerans_Agy99 KGYSATG--IDGSATAIERARDNAAKAGVSVN-FQVADATRLEG-LDNRF
MAB_0240|M.abscessus_ATCC_1997 AGCSVTG--VDLAPTAIARARNNTRTAGVEAQ-FVLGDATTLD--YEARF
Mflv_4471|M.gilvum_PYR-GCK AGYDVLG--VDGAPSAVEQARHNAKTRVVDAR-FDVADALDLGD--EPAY
Mvan_1888|M.vanbaalenii_PYR-1 AGYDVLG--IDGAPTAVEQARRNAEAQGVDAR-FELADALHLGP--DPTY
MSMEG_2067|M.smegmatis_MC2_155 LGYDVLG--IDFSPQAIEMARENARGRGVDAR-FAVGDAMALGDLGDGAY
TH_1722|M.thermoresistible__bu DGYTVVG--IDLSPTAIAAANRTAQERGLATASFVQGDITDFGG-YDGRF
MLBr_01723|M.leprae_Br4923 SGVARRGSVTDLSPGMVKIATRNGQSLGLDIN-GRVADAEGIPY-DDNTF
* * * : : * . : .* : : :
Mb2694c|M.bovis_AF2122/97 DTVVDCAFYHTFSTAPELQRCYVRALRRAS--------KPGARLYMFEFG
Rv2675c|M.tuberculosis_H37Rv DTVVDCAFYHTFSTAPELQRCYVRALRRAS--------KPGARLYMFEFG
MAV_3570|M.avium_104 DTVVDCAFYHVFATEPELRSSYARALHRAT--------KPGARLYMFEFG
MMAR_2041|M.marinum_M DTVVDCAFYHTFGTAPELQRSYVRALWRAT--------KPGARLYMYEFG
MUL_3309|M.ulcerans_Agy99 DTVVDCAFYHTFGTAPELQRSYVQALWRAT--------KPGARLYMYEFG
MAB_0240|M.abscessus_ATCC_1997 DTVVDSKLLDNLDSA-EARGRYLRSLYRAM--------KPQGRLFLYGFS
Mflv_4471|M.gilvum_PYR-GCK DTIIDSALFHIFDDA--DREKYVRSLGRAT--------APGSVVHLLALS
Mvan_1888|M.vanbaalenii_PYR-1 DTIVDSALFHIFDDA--DRATYVRSLHAAT--------RPGSVVHLLALS
MSMEG_2067|M.smegmatis_MC2_155 DTILDSALFHIFDDA--DRQTYVASLHAGC--------RPGGTVHILALS
TH_1722|M.thermoresistible__bu NTIIDSTLFHSIPVQ--ARDGYLRSIHRAA--------APGARYFALVFA
MLBr_01723|M.leprae_Br4923 DLVVGHAVLHHIPDVELSLREVLRVLKPGGRFVFAGEPTDVGNRYARVFA
: ::. . . : : . . . :.
Mb2694c|M.bovis_AF2122/97 EHNV---NGFSMPRSLSEDDFRQVLPVGGWEITYLGTTTYQVNLSVEALE
Rv2675c|M.tuberculosis_H37Rv EHNV---NGFSMPRSLSEDDFRQVLPVGGWEITYLGTTTYQVNLSVEALE
MAV_3570|M.avium_104 EHDV---NGFKMMRSLSENDFRDVLPAAGWEISYLGPTTYQVNLSAETIQ
MMAR_2041|M.marinum_M AHQV---NGLTAIRSLPEENFREMLPLGGWEITYLGPTTYRTTVSMETLE
MUL_3309|M.ulcerans_Agy99 AHQV---NGLTAIRSLPEENFREMLPLGGWEITYLGPTTYRTTVSMETLE
MAB_0240|M.abscessus_ATCC_1997 DGHV---NGFHN-HELDGIDYEMALTAAGFTITYLGETTYR--LSIPSYR
Mflv_4471|M.gilvum_PYR-GCK D------TGRGFGPEVSEETIRAAFADPGWDIEALTTTYYRGVITAPHAD
Mvan_1888|M.vanbaalenii_PYR-1 D------SGRGFGPEVSEHTIRAAFG-AGWEVEALTETTYRGVVIDAHTE
MSMEG_2067|M.smegmatis_MC2_155 D------AGRGFGPEVSEEQIRKAFG-DGWDLEALETTTYRGVVGPVHAE
TH_1722|M.thermoresistible__bu KGAFPEEMEQGPGPNPVDEDELREAVSKYWEIDEIRPAFIHANMPPDVPG
MLBr_01723|M.leprae_Br4923 DLTWKVTIRVVQLPGLSAWRRTQVEIDENSRAAALEWVVDLHTFEPKVLE
: . .
Mb2694c|M.bovis_AF2122/97 LMAARNPDMADQVRCVLERFRAI-------------------------KP
Rv2675c|M.tuberculosis_H37Rv LMAARNPDMADQVRCVLERFRAI-------------------------KP
MAV_3570|M.avium_104 MMAARNPDMADEAAKLLERFRAM-------------------------EP
MMAR_2041|M.marinum_M TMAARNPEVADQVGPLVERFKTI-------------------------QP
MUL_3309|M.ulcerans_Agy99 TMAARNPEVADQVGPLVERFKTI-------------------------QP
MAB_0240|M.abscessus_ATCC_1997 RICTDCP---DQVP------------------------------------
Mflv_4471|M.gilvum_PYR-GCK AMGLDVG-------------------------------------------
Mvan_1888|M.vanbaalenii_PYR-1 ALNLPAG-------------------------------------------
MSMEG_2067|M.smegmatis_MC2_155 AIGLPVG-------------------------------------------
TH_1722|M.thermoresistible__bu VESFDTDE------------------------------------------
MLBr_01723|M.leprae_Br4923 TMATNAGAVQVKTVTEEFTAAMLGWPLRTFESTMPPSKLGWGWARFAFTS
Mb2694c|M.bovis_AF2122/97 WLVGGRVHAPFWEVHATRVD------------
Rv2675c|M.tuberculosis_H37Rv WLVGGRVHAPFWEVHATRVD------------
MAV_3570|M.avium_104 WLQAGRVHAPFWEVHATRVD------------
MMAR_2041|M.marinum_M WLTDGIAHVPFWEVHATRLD------------
MUL_3309|M.ulcerans_Agy99 WLTDGIAHVPFWEVHATRPD------------
MAB_0240|M.abscessus_ATCC_1997 --PDGQIHIPMIEIHAVRNGLWQA--------
Mflv_4471|M.gilvum_PYR-GCK ----TVVDEPAWLARIRRL-------------
Mvan_1888|M.vanbaalenii_PYR-1 ----TVVDEPAWSARIRRL-------------
MSMEG_2067|M.smegmatis_MC2_155 ----TQVDEPAWLARARRL-------------
TH_1722|M.thermoresistible__bu ---KGRTKLPAFLLSAHKA-------------
MLBr_01723|M.leprae_Br4923 WKTLSWVDANVWRRVIPKSWFYNIIITGVKPS
. :