For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTVDLLSLSGRVVVVAGAAGGGIGTAVTRMVARAGATVVAVSRGQDNLDRQVGPLVAEGLAVLPVAADVC TDEGIEATLEAVRRADGDLHGLVNVAGGADPSTWMPSTRVRRSDWHELFARNLEAMFFMSQAIAAELKSR QRPGSIVSLSSISGMNAAPFHIGYGTAKAAIVAATRTMALELACDETAAGSPIRVNAVAPGVTETPASHT YTDVDPERDRRAIAMGRRGRPDEVAGVILFLLSDLSSYITGQTLLVDGGLNLKWGHLGADNTSLFLKDES FRAAIKRWDEA
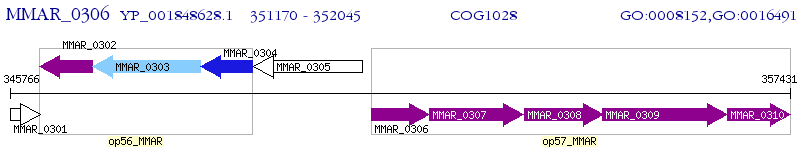
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0306 | - | - | 100% (291) | short-chain type dehydrogenase/reductase |
| M. marinum M | MMAR_4756 | - | 5e-35 | 37.45% (259) | oxidoreductase |
| M. marinum M | MMAR_4968 | - | 2e-26 | 32.45% (265) | short-chain type dehydrogenase/reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1963c | - | 5e-28 | 32.05% (259) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1787 | - | 1e-109 | 71.63% (282) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv1928c | - | 5e-28 | 32.05% (259) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 1e-19 | 28.52% (256) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_0296 | - | 6e-25 | 31.15% (260) | putative 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase |
| M. avium 104 | MAV_5208 | - | 1e-114 | 75.00% (276) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. smegmatis MC2 155 | MSMEG_5628 | - | 1e-116 | 76.51% (281) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. thermoresistible (build 8) | TH_1335 | - | 3e-72 | 77.97% (177) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. ulcerans Agy99 | MUL_0327 | - | 5e-34 | 37.07% (259) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_4963 | - | 1e-112 | 73.05% (282) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1963c|M.bovis_AF2122/97 -MSVLDLFDLHGKRALITGASTG-IGKRVAL-AYVEAGAQVAIAARHLDA
Rv1928c|M.tuberculosis_H37Rv -MSVLDLFDLHGKRALITGASTG-IGKRVAL-AYVEAGAQVAIAARHLDA
Mflv_1787|M.gilvum_PYR-GCK MT-TGVSINLDGRIVVVSGAGGGGIGTTVTR-LAADAGATVVAVSRSQDN
Mvan_4963|M.vanbaalenii_PYR-1 MSGTGISIRLDRRIVVVSGAGGGGIGTTVTR-LAAEAGATVVAVSRSKDN
MSMEG_5628|M.smegmatis_MC2_155 ---MDKLLNLDGRIVVVSGAGGGGIGTTVTR-MAAEAGATVVAVSRSQDN
TH_1335|M.thermoresistible__bu --------------------------------------------------
MAV_5208|M.avium_104 -------------MVVVSGAGGGGIGTTVTA-MAARAGATVIAVSRSKEN
MMAR_0306|M.marinum_M --MTVDLLSLSGRVVVVAGAAGGGIGTAVTR-MVARAGATVVAVSRGQDN
MUL_0327|M.ulcerans_Agy99 MAIDPADVLLNGRVAVVTGAGSG-IGRGVAA-GLAAFGASMAIWERDPQT
MLBr_01807|M.leprae_Br4923 ---------MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRL
MAB_0296|M.abscessus_ATCC_1997 ------------MVAVVTGAGGGVGSRVVT--TLARRHERVVAVDIDEVA
Mb1963c|M.bovis_AF2122/97 LEKLADEIGTSGGKVVP-----VCCDVSQHQQVTSMLDQVTAELGGIDIA
Rv1928c|M.tuberculosis_H37Rv LEKLADEIGTSGGKVVP-----VCCDVSQHQQVTSMLDQVTAELGGIDIA
Mflv_1787|M.gilvum_PYR-GCK LDEHVVPLAGEGRDVVT-----VAADASTDDGIATVLDTVRRTPGRLYGL
Mvan_4963|M.vanbaalenii_PYR-1 LDEHVVPLARKGLSVVP-----VAADASTDDGIATVLDHVRRTDGDLYGL
MSMEG_5628|M.smegmatis_MC2_155 LDTHVGPLADKGLSVVT-----VAADASTDDGIATVLDAVRRTDGRLYGL
TH_1335|M.thermoresistible__bu --------------------------------------------------
MAV_5208|M.avium_104 LDEHIAPLAAQGLAVLP-----VAADASTDEGIAAVIDQARRADGQLYGL
MMAR_0306|M.marinum_M LDRQVGPLVAEGLAVLP-----VAADVCTDEGIEATLEAVRRADGDLHGL
MUL_0327|M.ulcerans_Agy99 CAQAAAELGGLG----------IATDVRSGDQVDAALQRTQAELGAVTIL
MLBr_01807|M.leprae_Br4923 VADGHRVAVTHRASVVPDWFFGVECDVTDNDAIGAAFKAVEEHQGPVEVL
MAB_0296|M.abscessus_ATCC_1997 IKQLVEYAADEGLDVI-----GQILDITDDTAVAVGVSDIESRVGPITAL
Mb1963c|M.bovis_AF2122/97 VCNAG---IITVTPMLDMPLEEFQRLQNTNVTGVFLTAQAAAKAMVKQGQ
Rv1928c|M.tuberculosis_H37Rv VCNAG---IITVTPMLDMPLEEFQRLQNTNVTGVFLTAQAAAKAMVKQGQ
Mflv_1787|M.gilvum_PYR-GCK VNIAGGAAPATWMPSTRVTRDDWRALFQANLETAFFMSQAISGEIRSNGN
Mvan_4963|M.vanbaalenii_PYR-1 VNIAGGAAPATWMPSTRVTREDWRALFAANLETSFFMSQAVGSELRTRGL
MSMEG_5628|M.smegmatis_MC2_155 VNVAGGAAPSTWMPATRVSRADWRELFTANLETMFFMSQAVAAEIRDNGD
TH_1335|M.thermoresistible__bu -----------------VRRQDWRELFHRNLETAFFMSQAVANEIRAQGN
MAV_5208|M.avium_104 VNVAGGAEPSTWMPSTRVSRTDWRKIFADNLETAFFMSQAVAAELLARRL
MMAR_0306|M.marinum_M VNVAGGADPSTWMPSTRVRRSDWHELFARNLEAMFFMSQAIAAELKSRQR
MUL_0327|M.ulcerans_Agy99 VNNVGG---VFWSPLLQTSEKGWDALYRSNLGHVLLCTQRVARRLVAAGL
MLBr_01807|M.leprae_Br4923 VANAG---ITSDMLIMRMSEEQFQKVIDVNLTGVFRVAKLASRNMIKQR-
MAB_0296|M.abscessus_ATCC_1997 VNGAG---VLRTGAAVEFSDADWARTFDVNVRAPFVLSREVARRMIPRR-
: *: : :: . :
Mb1963c|M.bovis_AF2122/97 GGVIINTASMSGHIINVPQQVSHYCASKAAVIHLTKAMAVELAPHK----
Rv1928c|M.tuberculosis_H37Rv GGVIINTASMSGHIINVPQQVSHYCASKAAVIHLTKAMAVELAPHK----
Mflv_1787|M.gilvum_PYR-GCK PGAIVSVSSISG--MNTAPFHIAYGTAKAAIVAMTRTMAVELALDG----
Mvan_4963|M.vanbaalenii_PYR-1 RGSIVSVSSISG--MNTAPFHIAYGTAKAAIVAMTRTMAVELALDD----
MSMEG_5628|M.smegmatis_MC2_155 PGSIVSVSSISG--MNTAPFHIAYGTAKAAIVAATRTMAVELAAAG----
TH_1335|M.thermoresistible__bu PGSIVSISSISG--MNTAPFHIGYGTAKAAIVAMTRTMAVEHALHG----
MAV_5208|M.avium_104 PGSIVSISSISG--MNTAPFHIAYGTAKSAIAAMTRTMALELAQSA----
MMAR_0306|M.marinum_M PGSIVSLSSISG--MNAAPFHIGYGTAKAAIVAATRTMALELACDETAAG
MUL_0327|M.ulcerans_Agy99 PGSIMAITSIEG--VRAAPGYAAYAAAKAGVVNYTKTTALELAHHG----
MLBr_01807|M.leprae_Br4923 FGRYIFMGSVVG--LSGVPGQANYAASKSGLIGMARSLARELGSRS----
MAB_0296|M.abscessus_ATCC_1997 AGSVVTVASNAA--LVARKGMTAYAASKAAANAFTKSLGLELAEYG----
* : * . : * ::*:. ::: . * .
Mb1963c|M.bovis_AF2122/97 --IRVNSVSPGYILTELVEPY-TEYQPLWEP---------KIPLGRLGRP
Rv1928c|M.tuberculosis_H37Rv --IRVNSVSPGYILTELVEPY-TEYQPLWEP---------KIPLGRLGRP
Mflv_1787|M.gilvum_PYR-GCK --IRVNAVAPGVTETAASRMY-VDDDPERDRT--------AIAMGRRGTP
Mvan_4963|M.vanbaalenii_PYR-1 --IRVNAVAPGVTETAASRTY-VDEDPERDRT--------AIAMGRRGTP
MSMEG_5628|M.smegmatis_MC2_155 --IRVNAVAPGVTETAASATY-VDADPERDGR--------AIAMGRRGTP
TH_1335|M.thermoresistible__bu --IRVNAVAPGVTETAASRTY-VDEDPDRDRT--------AIAMGRRGRP
MAV_5208|M.avium_104 --IRVNAVAPGVTETAASRTY-VADDPDRDRR--------AIAMGRRGRP
MMAR_0306|M.marinum_M SPIRVNAVAPGVTETPASHTY-TDVDPERDRR--------AIAMGRRGRP
MUL_0327|M.ulcerans_Agy99 --IRVNAIAPDITMTEGLAQLGGAAALERIGS--------LVPLGRPGDV
MLBr_01807|M.leprae_Br4923 --ITANVVAPGFIDTEMTRAMDSKYQEAALQA---------IPLGRIGAV
MAB_0296|M.abscessus_ATCC_1997 --IRCNVVAPGSTDTPMLRAMLGDSSDAGLVAGSLGDYKPGIPLGRVAQP
* * ::*. * :.:** .
Mb1963c|M.bovis_AF2122/97 EELAGLYLYLASEASSYMTGSDIVIDGGYTCP------------------
Rv1928c|M.tuberculosis_H37Rv EELAGLYLYLASEASSYMTGSDIVIDGGYTCP------------------
Mflv_1787|M.gilvum_PYR-GCK EEQAGAILFLLSDLSGYVTGQTLLVDGGLNLKWSHLGADNTSLFLKDESF
Mvan_4963|M.vanbaalenii_PYR-1 EEQAGAILFLLSDLSSYITGQTLLVDGGLNLKWSHLAADNTSLFLKDESF
MSMEG_5628|M.smegmatis_MC2_155 DEQAGAILFLLSDLSNYVTGQTLLVDGGLNLRWTHLGADNTSLFLKDDNF
TH_1335|M.thermoresistible__bu EEQAGPILFLLSDLSSYITGQTLLVDGGLNLRWSHLGADNTSLFLRDESF
MAV_5208|M.avium_104 EEQAGAILFLLSELSSYVTGQTLLVDGGLDLRWSHLGADNTSLFLHDESF
MMAR_0306|M.marinum_M DEVAGVILFLLSDLSSYITGQTLLVDGGLNLKWGHLGADNTSLFLKDESF
MUL_0327|M.ulcerans_Agy99 DEIASVAVFLASDMARYVTGQTIHVDGGTHAAAGWYRDPQTGEYRLGPG-
MLBr_01807|M.leprae_Br4923 EEIAGAVSFLASEDAGYISGAVIPVDGGLGMGH-----------------
MAB_0296|M.abscessus_ATCC_1997 QHIADAVAFLLSDQAAHITMHTLVVDGGATLGV-----------------
:. *. :* *: : ::: : :***
Mb1963c|M.bovis_AF2122/97 ----------
Rv1928c|M.tuberculosis_H37Rv ----------
Mflv_1787|M.gilvum_PYR-GCK REVISRH---
Mvan_4963|M.vanbaalenii_PYR-1 RESIRRI---
MSMEG_5628|M.smegmatis_MC2_155 RAAITQP---
TH_1335|M.thermoresistible__bu RAAIKEM---
MAV_5208|M.avium_104 RAAIRRM---
MMAR_0306|M.marinum_M RAAIKRWDEA
MUL_0327|M.ulcerans_Agy99 ----------
MLBr_01807|M.leprae_Br4923 ----------
MAB_0296|M.abscessus_ATCC_1997 ----------