For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVVSGAGGGGIGTTVTAMAARAGATVIAVSRSKENLDEHIAPLAAQGLAVLPVAADASTDEGIAAVIDQA RRADGQLYGLVNVAGGAEPSTWMPSTRVSRTDWRKIFADNLETAFFMSQAVAAELLARRLPGSIVSISSI SGMNTAPFHIAYGTAKSAIAAMTRTMALELAQSAIRVNAVAPGVTETAASRTYVADDPDRDRRAIAMGRR GRPEEQAGAILFLLSELSSYVTGQTLLVDGGLDLRWSHLGADNTSLFLHDESFRAAIRRM*
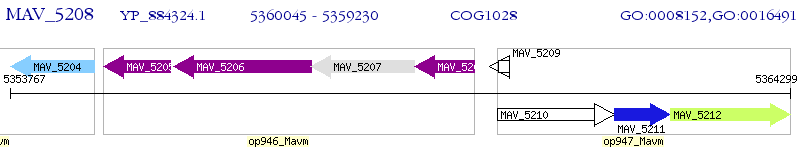
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_5208 | - | - | 100% (271) | oxidoreductase, short chain dehydrogenase/reductase family protein |
| M. avium 104 | MAV_0894 | - | 3e-29 | 34.52% (252) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. avium 104 | MAV_3012 | - | 7e-29 | 33.47% (248) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0950c | - | 1e-31 | 38.37% (245) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1787 | - | 1e-117 | 77.04% (270) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv0927c | - | 1e-31 | 38.37% (245) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 2e-20 | 28.28% (244) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_0296 | - | 3e-30 | 32.68% (254) | putative 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase |
| M. marinum M | MMAR_0306 | - | 1e-114 | 75.00% (276) | short-chain type dehydrogenase/reductase |
| M. smegmatis MC2 155 | MSMEG_5628 | - | 1e-117 | 78.73% (268) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. thermoresistible (build 8) | TH_1335 | - | 5e-76 | 82.66% (173) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. ulcerans Agy99 | MUL_0327 | - | 2e-33 | 37.45% (243) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_4963 | - | 1e-122 | 79.70% (271) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0950c|M.bovis_AF2122/97 --MILDMFRLDDKVAVITG-GGRGLGAAIALAFAQAGADVLIASRTSSEL
Rv0927c|M.tuberculosis_H37Rv --MILDMFRLDDKVAVITG-GGRGLGAAIALAFAQAGADVLIASRTSSEL
Mflv_1787|M.gilvum_PYR-GCK MT-TGVSINLDGRIVVVSGAGGGGIGTTVTRLAADAGATVVAVSRSQDNL
Mvan_4963|M.vanbaalenii_PYR-1 MSGTGISIRLDRRIVVVSGAGGGGIGTTVTRLAAEAGATVVAVSRSKDNL
MSMEG_5628|M.smegmatis_MC2_155 ---MDKLLNLDGRIVVVSGAGGGGIGTTVTRMAAEAGATVVAVSRSQDNL
TH_1335|M.thermoresistible__bu --------------------------------------------------
MAV_5208|M.avium_104 -------------MVVVSGAGGGGIGTTVTAMAARAGATVIAVSRSKENL
MMAR_0306|M.marinum_M --MTVDLLSLSGRVVVVAGAAGGGIGTAVTRMVARAGATVVAVSRGQDNL
MUL_0327|M.ulcerans_Agy99 MAIDPADVLLNGRVAVVTG-AGSGIGRGVAAGLAAFGASMAIWERDPQTC
MLBr_01807|M.leprae_Br4923 ---------MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRL
MAB_0296|M.abscessus_ATCC_1997 ------------MVAVVTGAGGGVGSRVVT--TLARRHERVVAVDIDEVA
Mb0950c|M.bovis_AF2122/97 DAVAEQIRAAGRRAHT------VAADLAHPEVTAQLAGQAVGAFGKLDIV
Rv0927c|M.tuberculosis_H37Rv DAVAEQIRAAGRRAHT------VAADLAHPEVTAQLAGQAVGAFGKLDIV
Mflv_1787|M.gilvum_PYR-GCK DEHVVPLAGEGRDVVT------VAADASTDDGIATVLDTVRRTPGRLYGL
Mvan_4963|M.vanbaalenii_PYR-1 DEHVVPLARKGLSVVP------VAADASTDDGIATVLDHVRRTDGDLYGL
MSMEG_5628|M.smegmatis_MC2_155 DTHVGPLADKGLSVVT------VAADASTDDGIATVLDAVRRTDGRLYGL
TH_1335|M.thermoresistible__bu --------------------------------------------------
MAV_5208|M.avium_104 DEHIAPLAAQGLAVLP------VAADASTDEGIAAVIDQARRADGQLYGL
MMAR_0306|M.marinum_M DRQVGPLVAEGLAVLP------VAADVCTDEGIEATLEAVRRADGDLHGL
MUL_0327|M.ulcerans_Agy99 AQAAAELGGLG-----------IATDVRSGDQVDAALQRTQAELGAVTIL
MLBr_01807|M.leprae_Br4923 VADGHRVAVTHRASVVPDWFFGVECDVTDNDAIGAAFKAVEEHQGPVEVL
MAB_0296|M.abscessus_ATCC_1997 IKQLVEYAADEGLDVI-----GQILDITDDTAVAVGVSDIESRVGPITAL
Mb0950c|M.bovis_AF2122/97 VNNVGGTMPNTLLSTS---TKDLADAFAFNVGTAHALTVAAVPLMLEHSG
Rv0927c|M.tuberculosis_H37Rv VNNVGGTMPNTLLSTS---TKDLADAFAFNVGTAHALTVAAVPLMLEHSG
Mflv_1787|M.gilvum_PYR-GCK VNIAGGAAPATWMPSTRVTRDDWRALFQANLETAFFMSQAISGEIRSNGN
Mvan_4963|M.vanbaalenii_PYR-1 VNIAGGAAPATWMPSTRVTREDWRALFAANLETSFFMSQAVGSELRTRGL
MSMEG_5628|M.smegmatis_MC2_155 VNVAGGAAPSTWMPATRVSRADWRELFTANLETMFFMSQAVAAEIRDNGD
TH_1335|M.thermoresistible__bu -----------------VRRQDWRELFHRNLETAFFMSQAVANEIRAQGN
MAV_5208|M.avium_104 VNVAGGAEPSTWMPSTRVSRTDWRKIFADNLETAFFMSQAVAAELLARRL
MMAR_0306|M.marinum_M VNVAGGADPSTWMPSTRVRRSDWHELFARNLEAMFFMSQAIAAELKSRQR
MUL_0327|M.ulcerans_Agy99 VNNVGGVFWSPLLQTS---EKGWDALYRSNLGHVLLCTQRVARRLVAAGL
MLBr_01807|M.leprae_Br4923 VANAGITSDMLIMRMS---EEQFQKVIDVNLTGVFRVAKLASRNMIKQR-
MAB_0296|M.abscessus_ATCC_1997 VNGAGVLRTGAAVEFS---DADWARTFDVNVRAPFVLSREVARRMIPRR-
*: : :
Mb0950c|M.bovis_AF2122/97 GGSVINISSTMGRLAARGFAAYGTAKAALAHYTRLAALDLCPRV------
Rv0927c|M.tuberculosis_H37Rv GGSVINISSTMGRLAARGFAAYGTAKAALAHYTRLAALDLCPRV------
Mflv_1787|M.gilvum_PYR-GCK PGAIVSVSSISGMNTAPFHIAYGTAKAAIVAMTRTMAVELALDG------
Mvan_4963|M.vanbaalenii_PYR-1 RGSIVSVSSISGMNTAPFHIAYGTAKAAIVAMTRTMAVELALDD------
MSMEG_5628|M.smegmatis_MC2_155 PGSIVSVSSISGMNTAPFHIAYGTAKAAIVAATRTMAVELAAAG------
TH_1335|M.thermoresistible__bu PGSIVSISSISGMNTAPFHIGYGTAKAAIVAMTRTMAVEHALHG------
MAV_5208|M.avium_104 PGSIVSISSISGMNTAPFHIAYGTAKSAIAAMTRTMALELAQSA------
MMAR_0306|M.marinum_M PGSIVSLSSISGMNAAPFHIGYGTAKAAIVAATRTMALELACDETAAGSP
MUL_0327|M.ulcerans_Agy99 PGSIMAITSIEGVRAAPGYAAYAAAKAGVVNYTKTTALELAHHG------
MLBr_01807|M.leprae_Br4923 FGRYIFMGSVVGLSGVPGQANYAASKSGLIGMARSLARELGSRS------
MAB_0296|M.abscessus_ATCC_1997 AGSVVTVASNAALVARKGMTAYAASKAAANAFTKSLGLELAEYG------
* : : * . *.::*:. :: . :
Mb0950c|M.bovis_AF2122/97 -RVNAIAPGSILTSALEVVAA---NDELRAP----MEQATPLRRLGDPVD
Rv0927c|M.tuberculosis_H37Rv -RVNAIAPGSILTSALEVVAA---NDELRAP----MEQATPLRRLGDPVD
Mflv_1787|M.gilvum_PYR-GCK IRVNAVAPGVTETAASRMYVD---DDPERD------RTAIAMGRRGTPEE
Mvan_4963|M.vanbaalenii_PYR-1 IRVNAVAPGVTETAASRTYVD---EDPERD------RTAIAMGRRGTPEE
MSMEG_5628|M.smegmatis_MC2_155 IRVNAVAPGVTETAASATYVD---ADPERD------GRAIAMGRRGTPDE
TH_1335|M.thermoresistible__bu IRVNAVAPGVTETAASRTYVD---EDPDRD------RTAIAMGRRGRPEE
MAV_5208|M.avium_104 IRVNAVAPGVTETAASRTYVA---DDPDRD------RRAIAMGRRGRPEE
MMAR_0306|M.marinum_M IRVNAVAPGVTETPASHTYTD---VDPERD------RRAIAMGRRGRPDE
MUL_0327|M.ulcerans_Agy99 IRVNAIAPDITMTEGLAQLGG---AAALER-----IGSLVPLGRPGDVDE
MLBr_01807|M.leprae_Br4923 ITANVVAPGFIDTEMTRAMDSKYQEAALQA---------IPLGRIGAVEE
MAB_0296|M.abscessus_ATCC_1997 IRCNVVAPGSTDTPMLRAMLGDSSDAGLVAGSLGDYKPGIPLGRVAQPQH
*.:**. * .: * . .
Mb0950c|M.bovis_AF2122/97 IAAAAVYLASPAGSFLTGKTLEVDGGLTFPNLDLPIPDL-----------
Rv0927c|M.tuberculosis_H37Rv IAAAAVYLASPAGSFLTGKTLEVDGGLTFPNLDLPIPDL-----------
Mflv_1787|M.gilvum_PYR-GCK QAGAILFLLSDLSGYVTGQTLLVDGGLNLKWSHLGADNTSLFLKDESFRE
Mvan_4963|M.vanbaalenii_PYR-1 QAGAILFLLSDLSSYITGQTLLVDGGLNLKWSHLAADNTSLFLKDESFRE
MSMEG_5628|M.smegmatis_MC2_155 QAGAILFLLSDLSNYVTGQTLLVDGGLNLRWTHLGADNTSLFLKDDNFRA
TH_1335|M.thermoresistible__bu QAGPILFLLSDLSSYITGQTLLVDGGLNLRWSHLGADNTSLFLRDESFRA
MAV_5208|M.avium_104 QAGAILFLLSELSSYVTGQTLLVDGGLDLRWSHLGADNTSLFLHDESFRA
MMAR_0306|M.marinum_M VAGVILFLLSDLSSYITGQTLLVDGGLNLKWGHLGADNTSLFLKDESFRA
MUL_0327|M.ulcerans_Agy99 IASVAVFLASDMARYVTGQTIHVDGGTHAAAGWYRDPQTGEYRLGPG---
MLBr_01807|M.leprae_Br4923 IAGAVSFLASEDAGYISGAVIPVDGGLGMGH-------------------
MAB_0296|M.abscessus_ATCC_1997 IADAVAFLLSDQAAHITMHTLVVDGGATLGV-------------------
* :* * . .:: .: ****
Mb0950c|M.bovis_AF2122/97 --------
Rv0927c|M.tuberculosis_H37Rv --------
Mflv_1787|M.gilvum_PYR-GCK VISRH---
Mvan_4963|M.vanbaalenii_PYR-1 SIRRI---
MSMEG_5628|M.smegmatis_MC2_155 AITQP---
TH_1335|M.thermoresistible__bu AIKEM---
MAV_5208|M.avium_104 AIRRM---
MMAR_0306|M.marinum_M AIKRWDEA
MUL_0327|M.ulcerans_Agy99 --------
MLBr_01807|M.leprae_Br4923 --------
MAB_0296|M.abscessus_ATCC_1997 --------