For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTSGSAPQWLTRNLRVLSLVSLLQDAASELLYPLLPIYLTTVLGAPPSVVGAVEGAAEGAASLTKVVSGP LGDRFARRPLIASGYGMAALGKLIVAAAGAWPGVLAGRVVDRLGKGLRGAPRDALLVDGVEPAQRGRAFG FHRTMDTLGAVIGPLLGLAGYELFDHQIGPVLYLAVIPAVLSVLLITWVREPPRNLAKAKPASILDGTRR LSRRYWAVVSFLVLFSIANFPDALLLLRLKQIGFSVVGVIMAYVGYNAVYALASFPAGALADRLGRPVVY AFGLVFFAVGYLGLGITTDTATAWALIAAYGLFTACYDGVGKAWISAIVSAELQSSAQGVFQGLSGFAVL AAGLWAGFLWGVDGRLPLLISGGAGAVFALALLAAAAGTRRRAR
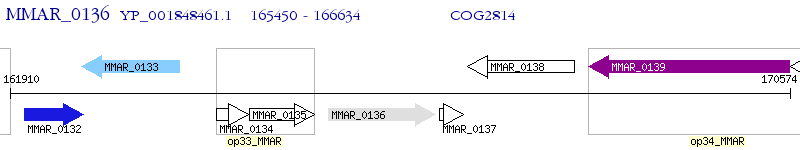
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0136 | - | - | 100% (394) | intergral membrane protein |
| M. marinum M | MMAR_5094 | - | 4e-11 | 23.98% (392) | integral membrane transport protein |
| M. marinum M | MMAR_4767 | - | 2e-07 | 28.78% (205) | integral membrane transport protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2288 | - | 1e-22 | 26.04% (361) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_1475 | - | 1e-09 | 26.06% (307) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv2265 | - | 1e-22 | 26.04% (361) | integral membrane protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4615 | - | 1e-24 | 26.45% (397) | major facilitator transporter |
| M. avium 104 | MAV_1539 | - | 6e-13 | 26.74% (359) | major facilitator family protein transporter |
| M. smegmatis MC2 155 | MSMEG_2380 | - | 5e-07 | 24.61% (321) | sugar transporter family protein |
| M. thermoresistible (build 8) | TH_0476 | - | 2e-09 | 23.75% (341) | major facilitator superfamily protein MFS_1 |
| M. ulcerans Agy99 | MUL_4961 | - | 0.0 | 97.97% (394) | intergral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_2778 | - | 1e-30 | 29.72% (397) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0136|M.marinum_M -------------MTSGSAPQWLTRNLRVLSLVSLLQDAASELLYP---L
MUL_4961|M.ulcerans_Agy99 -------------MTSGSAPQWLTRNLRVLSLVSLLQDAASELLYP---L
Mb2288|M.bovis_AF2122/97 --MGANGDVALSRIGATRPALSAWRFVTVFGVVGLLADVVYEGARS---I
Rv2265|M.tuberculosis_H37Rv --MGANGDVALSRIGATRPALSAWRFVTVFGVVGLLADVVYEGARS---I
MAB_4615|M.abscessus_ATCC_1997 ----------------MPQRLSAWRFVTTFGVISMLADIVYEGARS---I
Mvan_2778|M.vanbaalenii_PYR-1 ------------MTGTPGVGLSAWRFVTTFGVVSLLADIVYEGARS---I
Mflv_1475|M.gilvum_PYR-GCK -----------------MAPLSPTAQRAPLLAAGFTTAFGAHAVAG---T
TH_0476|M.thermoresistible__bu ----------------------VRRFAFPLLAYAFAAVMVGTTVPT---P
MAV_1539|M.avium_104 -------MVMPNGPVAHQAGTRGYRRMTAALCGAGLASFAAMYCTQ---A
MSMEG_2380|M.smegmatis_MC2_155 MELRFDILRSRDSMERMAAPVIGTLRRWSMLVIALTSTMCANVFINGAAF
.
MMAR_0136|M.marinum_M LPIYLTTVLGAPPSVVGAVEGAAEGAASLTKVVSGPLGDRFAR-RPLIAS
MUL_4961|M.ulcerans_Agy99 LPIYLATVLGAPPSVVGAVEGAAEGAASLTKIVSGPLGDRFAR-RPLIAS
Mb2288|M.bovis_AF2122/97 TGPLLAS-LGATGLVVGVVTGVGEAAALGLRLVSGPLADRSRRFWAWTIA
Rv2265|M.tuberculosis_H37Rv TGPLLAS-LGATGLVVGVVTGVGEAAALGLRLVSGPLADRSRRFWAWTIA
MAB_4615|M.abscessus_ATCC_1997 TGPLLAS-LGASALVVGVVTGIGEAAALALRLVSGPLTDRTRKFWAWTIS
Mvan_2778|M.vanbaalenii_PYR-1 TGPLLAS-LGATAVVVGVVTGVGEAAALILRMVSGPLADRTGRFWTWTIG
Mflv_1475|M.gilvum_PYR-GCK LGTTTTG-TAASLLTLGAMLAIYDGAEVILKPLFGTLADRIGA-RTVLIA
TH_0476|M.thermoresistible__bu MYAVYAERLGFGVLTTTVVFAGYTAGVLFALVVFGRWSDALGR-RPMLLA
MAV_1539|M.avium_104 LLPALSAHHRIGPATAALTVSLTTGALALSIIPASVLSERYGRTRVMLIS
MSMEG_2380|M.smegmatis_MC2_155 LIPTLHAERGLDLARSGLLSSMPSFGMVTTLIAWGYVVDRFGE-RLVLGL
. . . :
MMAR_0136|M.marinum_M GYGMAALGKLIVAAAGAWPGVLAGRVVDRLGKGLRGAPRDALLVDGVEPA
MUL_4961|M.ulcerans_Agy99 GYGMAALGKLIVAAAGAWPGVLAGRVVDRLGKGLRGAPRDALLVDGVDPA
Mb2288|M.bovis_AF2122/97 GYTLTVVTVPLLGIAGALWVACALVIAERVGKAVRGPAKDTLLSHAASVT
Rv2265|M.tuberculosis_H37Rv GYTLTVVTVPLLGIAGALWVACALVIAERVGKAVRGPAKDTLLSHAASVT
MAB_4615|M.abscessus_ATCC_1997 GYALTVVTVPFLGVSSVLWIAATLVIAERVGKAVRSPAKDTLLSHATAVT
Mvan_2778|M.vanbaalenii_PYR-1 GYFLTVVTVPFLGAVGVLWAAAALIIAERVGKAVRSPAKDTLLSHATAVT
Mflv_1475|M.gilvum_PYR-GCK GLAVFAVASLSFAVADETAWLWAARLGQGIGAAAFSPAASALVARLAPAS
TH_0476|M.thermoresistible__bu GVGCAIVSALVFLIAGSVPQLIAARFVSGMSAGIFTGTATAAVIEAAPER
MAV_1539|M.avium_104 GVASSVIG-LLLPFSPSLGVLVFGRAVQGVALAGIPAVAMALLAEEVDAS
MSMEG_2380|M.smegmatis_MC2_155 GSALTAAAALAAASVQSLFAVGLFLLLGGMAAASSNSASGRLVVGWFPPE
* :. . :
MMAR_0136|M.marinum_M QRGRAFGFHRTMDTLGAVIGPLLGLAGYELFDHQIGPVLYLAVIPAVLSV
MUL_4961|M.ulcerans_Agy99 QRGRAFGFHRTMDTLGAVIGPLLGLAGYELFDHQIGPVLYLAVIPAMLSV
Mb2288|M.bovis_AF2122/97 GRGRGFAVHEALDQVGAMIGPLTVAGMLAITGNAYAPALGVLTLPGGAAL
Rv2265|M.tuberculosis_H37Rv GRGRGFAVHEALDQVGAMIGPLTVAGMLAITGNAYAPALGVLTLPGGAAL
MAB_4615|M.abscessus_ATCC_1997 GRGKGFAVHEAMDQIGAITGPLLVAGMLALTHVNYLPTFAILALPGGATL
Mvan_2778|M.vanbaalenii_PYR-1 GRGRGFAVHEAMDQIGAVAGPLAVAAMLVVFGGDYAPTLALLAVPGVGVL
Mflv_1475|M.gilvum_PYR-GCK GQGRAFGTYGSYKSVGYTLGPLIGGAIVTFGG-----LQALFAVMAALAA
TH_0476|M.thermoresistible__bu RRSRAAAVATVANIGGLGAGPLLAGILVQYAPWPLHLTFWVHIVLMVLAA
MAV_1539|M.avium_104 SLGSAMGRYIAGTTIGGLAGRIVPSVVVQVGTWRVALLACSLITLAGTAV
MSMEG_2380|M.smegmatis_MC2_155 QRGLVMGIRQTATPLGVGLGALVIPRLAESHGVSAALLFPAVVCGIAAVV
. . * * :
MMAR_0136|M.marinum_M LLITWVR----EPPRN----LAKAKPASILDGTRRLSRRYWAVVSFLVLF
MUL_4961|M.ulcerans_Agy99 LLIAWVR----EPPRN----LAKAKPASILDGTRRLPRRYWAVVSFLVLF
Mb2288|M.bovis_AF2122/97 ALLLWLQRRVPRPESY--EDCPVVLGNPSAPRPWALPAQFWLYCGFTAIT
Rv2265|M.tuberculosis_H37Rv ALLLWLQRRVPRPESY--EDCPVVLGNPSAPRPWALPAQFWLYCGFTAIT
MAB_4615|M.abscessus_ATCC_1997 LLLFWLRRRVPHPERY--EHDDYIRGAADRPHKLSLPLSFWLYSGFTAIT
Mvan_2778|M.vanbaalenii_PYR-1 ALLAWLRVRVPEPARYEVEAAPSTKPVARVSFLRSLPRSFWTYAGFSAAT
Mflv_1475|M.gilvum_PYR-GCK AVAVWAAVAVPRLAVLP-RTRQTLLDTVRRFSESSFVTPTLALAAATAAL
TH_0476|M.thermoresistible__bu AAIAVAPETSPRTGRLG-------VQRLSLPPQVRKVFAIASLAAFAGFA
MAV_1539|M.avium_104 FAVLVPRSRFFTPKPAS-------VRAALRNLAGHLRNPVLAKLFAVGFV
MSMEG_2380|M.smegmatis_MC2_155 SAVFVLDPPRPPRAEAP---------AEHLANPYRGSSVLWRIHAVSVLL
MMAR_0136|M.marinum_M SIA--NFPDALLLLRLKQIG-FSVVGVIMAYVGYNAVYALASFPAGALAD
MUL_4961|M.ulcerans_Agy99 SIA--NFPDALLLLRLKQIG-FSVVGVIMAYVGYNAVYALASFPVGALAD
Mb2288|M.bovis_AF2122/97 MLG--FGTFGLLSFHMVSHGVLAAAMVPVVYAAAMAADALTALASGFSYD
Rv2265|M.tuberculosis_H37Rv MLG--FGTFGLLSFHMVSHGVLAAAMVPVVYAAAMAADALTALASGFSYD
MAB_4615|M.abscessus_ATCC_1997 MIG--FATFGVLSFHMVQRGVLPVPAVPIVYAAAMAADAVAALLSGWAYD
Mvan_2778|M.vanbaalenii_PYR-1 MLG--FTTFGVLSFHQVERGVVHVAVVPVIYAAAMVADALAALATGWVYD
Mflv_1475|M.gilvum_PYR-GCK SAG--VGFLPVLGTDFGLSPVVTGAVVSVLALTTAVVQPLAGRAADSGRI
TH_0476|M.thermoresistible__bu VTG--LFTAVAPSFLAEVIGVRNHAVAGAVAASIFAASAVTQVLGASVSP
MAV_1539|M.avium_104 LMGGFVTVYNYLGYRLAARPFGLAPSVVGLLFLLYLVGTGTSVVAGRLAD
MSMEG_2380|M.smegmatis_MC2_155 VVPQGVLWTFALVWLITDRG-WSAASAGILVTVTQLLGAGGRIAAGKWSD
. .
MMAR_0136|M.marinum_M RLGR---PVVYAFGLVFFAVGYLGLGITTDTATAWALIAAYGLFTACYDG
MUL_4961|M.ulcerans_Agy99 RLGR---PVVYAFGLVLFAVGYLGLGITTDTATAWALIAAYGLFTACYDG
Mb2288|M.bovis_AF2122/97 RYGA---KTLAVLPILSILVVLFAF--TDNVTMVVIGTLVWGAAVGIQES
Rv2265|M.tuberculosis_H37Rv RYGA---KTLAVLPILSILVVLFAF--TDNVTMVVIGTLVWGAAVGIQES
MAB_4615|M.abscessus_ATCC_1997 RVGP---KSLAALPIVGATVPMLAF--TDSLPGIVLGALLWGAAVGIQES
Mvan_2778|M.vanbaalenii_PYR-1 KVGA---VVLGALPVMAGLVPALAF--NDHLVPVVAGALVWGAAVGIQES
Mflv_1475|M.gilvum_PYR-GCK SLTS---GLFTGVVVTAAGVAAAALP---GLVGLLGGAVVIGVGCGLITP
TH_0476|M.thermoresistible__bu RRAV---AVGCALLIVGMVFLAAALV-WSSLPALIATAVVAGAGQGLSFS
MAV_1539|M.avium_104 RRGR---PLVLGAALP-IAVAGLLLTVPPSLAAIVAGVGVFTGGFFAAHT
MSMEG_2380|M.smegmatis_MC2_155 VIGYRLRPIRTIALAAAVTMAALALTDWLSSPVSIALMVAASVITVSDNG
. :
MMAR_0136|M.marinum_M VGKAWISAIVSAELQ---SSAQGVFQGLSGFAVLAAGLWAGFLWGVDGRL
MUL_4961|M.ulcerans_Agy99 VGKAWISAIVSAELQ---SSAQGVFQGLSGFAVLAAGLWAGFLWGVDGRL
Mb2288|M.bovis_AF2122/97 TLRGVVADLVASPRR---ASAYGVFAAGLGAATAGGGALIGWLYDIS--I
Rv2265|M.tuberculosis_H37Rv TLRGVVADLVASPRR---ASAYGVFAAGLGAATAGGGALIGWLYDIS--I
MAB_4615|M.abscessus_ATCC_1997 TLRAVVADLVPPPRR---ATAYGVYAAVLGTATAIGGALTGYLYGVS--I
Mvan_2778|M.vanbaalenii_PYR-1 TLRAVVADLVEPARR---ATAYGIYAAVLGCAAAAGGALTGYLYGQS--V
Mflv_1475|M.gilvum_PYR-GCK LAFTMLARSTPEERL---GQTMGAAEIGRECGDAAGPLVVAAIAAAS-TV
TH_0476|M.thermoresistible__bu RGLAAVSARTPSDQRGEVSSTYFVVAYVALLVPVIGEGWAAHHWGLR-TA
MAV_1539|M.avium_104 VASGWVGAVAQRDRA----EASALYLFSYYLGGSVAGAFGGVLYGVGGWS
MSMEG_2380|M.smegmatis_MC2_155 LAFTGIAEIAGPFWS---GRALGTQNTSQHLATAVSSPVFGGLIGVAG-Y
:. . : . .
MMAR_0136|M.marinum_M PLLISGGAGAVFALALLAAAAGTRRRAR------------
MUL_4961|M.ulcerans_Agy99 PLLISGGAGAVFALALLAAAAGTRRRAR------------
Mb2288|M.bovis_AF2122/97 GTLVVVVIALELMALVMMFAIRLPRVAPS-----------
Rv2265|M.tuberculosis_H37Rv GTLVVVVIALELMALVMMFAIRLPRVAPS-----------
MAB_4615|M.abscessus_ATCC_1997 PMLIAVVGVIQLVALITAAPTLLRHAGQV-----------
Mvan_2778|M.vanbaalenii_PYR-1 PLLVGVVIGVQAIALAVLPVVR--RVAR------------
Mflv_1475|M.gilvum_PYR-GCK PFGFLGLAAVVAASGVGVRRGRGRVGGVG-----------
TH_0476|M.thermoresistible__bu GISFAVGVAVLAIGCLIAILIQESRDPQAI----------
MAV_1539|M.avium_104 ATVCFVVVLLMAGAALVALLVRDNGFRIGRRVVAGVASVK
MSMEG_2380|M.smegmatis_MC2_155 PVAFAVCALLPALAIPLVPVEPARES--------------