For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VDVNRPDYVLGHSDYELGRLGRQAMLLDATTREYLATAGLEPGMRVLDVGSGPGDVAFLASELTMPGGEV VGTDRAAGAVSAASAAAAARGLSETVRFRLGDPADSSFERPFDAIVGRYVLTFQADPAAMLHRLSQHLVP GGIVAFHEPDWSAVRSSPPAPTHDRCCHWIREALDRTHTSWDMADRLHNAFTGAGLPAPTLTMRTFIGAG RAAAVWLQALADLVETLLPTIQAQGVATATEVGIETLTQRLLEEVSDLGATVIGRSEVGAWTRTPRSG
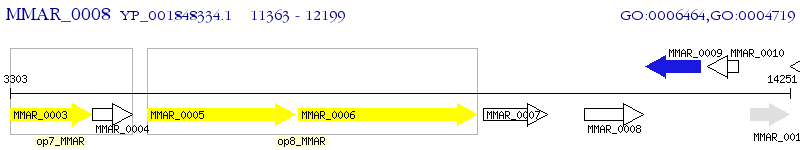
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0008 | - | - | 100% (278) | hypothetical protein MMAR_0008 |
| M. marinum M | MMAR_4513 | - | 9e-10 | 36.13% (119) | hypothetical protein MMAR_4513 |
| M. marinum M | MMAR_4799 | - | 5e-08 | 29.24% (171) | hypothetical protein MMAR_4799 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0862 | - | 3e-13 | 30.43% (207) | hypothetical protein Mb0862 |
| M. gilvum PYR-GCK | Mflv_1845 | - | 1e-07 | 34.86% (109) | methyltransferase type 11 |
| M. tuberculosis H37Rv | Rv0839 | - | 3e-13 | 30.43% (207) | hypothetical protein Rv0839 |
| M. leprae Br4923 | MLBr_00551 | - | 1e-06 | 26.32% (247) | putative methyltransferase |
| M. abscessus ATCC 19977 | MAB_4742c | - | 2e-06 | 32.08% (106) | hypothetical protein MAB_4742c |
| M. avium 104 | MAV_0854 | - | 9e-13 | 34.76% (164) | methyltransferase-UbiE family protein |
| M. smegmatis MC2 155 | MSMEG_0098 | - | 9e-08 | 29.93% (147) | methyltransferase |
| M. thermoresistible (build 8) | TH_3050 | - | 1e-11 | 41.82% (110) | PUTATIVE Methyltransferase type 12 |
| M. ulcerans Agy99 | MUL_0010 | - | 1e-153 | 96.40% (278) | hypothetical protein MUL_0010 |
| M. vanbaalenii PYR-1 | Mvan_4620 | - | 1e-07 | 28.34% (187) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1845|M.gilvum_PYR-GCK --------------MATYDVPDAFDAGADAYDGLVGANPGY---------
TH_3050|M.thermoresistible__bu -------VTGRRRHLDRAAVPAAFDAGAASYDRLVAANPGY---------
MMAR_0008|M.marinum_M ----------------MDVNRPDYVLGHSDYELGRLG-------------
MUL_0010|M.ulcerans_Agy99 ----------------MNVNRPDYVLGHSDYELGRLG-------------
Mb0862|M.bovis_AF2122/97 ---------------MNDKRRAIYTHGYHESVLRSHR-------------
Rv0839|M.tuberculosis_H37Rv ---------------MNDKRRAIYTHGYHESVLRSHR-------------
MAV_0854|M.avium_104 ---------------MNDQHRATYTHGHHESVLRSHR-------------
Mvan_4620|M.vanbaalenii_PYR-1 ---------------------MSFVVASEAYGRFMGR-------------
MSMEG_0098|M.smegmatis_MC2_155 ---MDTALRRAMDLLIDPPAEPDFSKGYLDLLGRGDSTAPKNTGLIQKAW
MLBr_00551|M.leprae_Br4923 -------------MNLYTPIHEDQSLAAIHRAMWALG-------------
MAB_4742c|M.abscessus_ATCC_199 MTAPSANPVPETTEEFSERLTAAIDNAALVLLMSIGHQTGLLDTLAASAG
.
Mflv_1845|M.gilvum_PYR-GCK -------------HKHLRLSARRMGLP---DGGRGLVLLDIGCGTG-AST
TH_3050|M.thermoresistible__bu -------------HEHLRLSAHRMGLPGCAARXRGLRILDAGCGTG-AST
MMAR_0008|M.marinum_M -------------RQAMLLDATTREYLATAGLEPGMRVLDVGSGPGDVAF
MUL_0010|M.ulcerans_Agy99 -------------RQAMLLDATTREYLATAGLEPGMRVLDVGSGPGDVAF
Mb0862|M.bovis_AF2122/97 -------------RRTAENSAG---YLLPY-LVPGLSVLDVGCGPGTITV
Rv0839|M.tuberculosis_H37Rv -------------RRTAENSAG---YLLPY-LVPGLSVLDVGCGPGTITV
MAV_0854|M.avium_104 -------------RRTAEDSAG---YLLAH-LTPGLSVLDVGCGPGTITA
Mvan_4620|M.vanbaalenii_PYR-1 -----------------YAEPLAEAFVAFAGVGVGDKVLDVGCGPGALTA
MSMEG_0098|M.smegmatis_MC2_155 ASPLGSKLYDRAQLLNRRLVASTRPPVEWLRIPPGGTVLDIGCGPGNITA
MLBr_00551|M.leprae_Br4923 ---------DYALMAEEVMAPLGPILVTATGIRPGMQVLDVAAGSGNISL
MAB_4742c|M.abscessus_ATCC_199 STSEQLADAAGLEERYVREWLAGMTVAHVVDYEPVSAIYTLPSHRAATLT
: . .
Mflv_1845|M.gilvum_PYR-GCK AALLRAAPHAEIVAVDGSAGMLAQARAKRWPPNVRF-VHSRVEDLAVNGV
TH_3050|M.thermoresistible__bu AALLTYAPDAEIVAVDASAGMLAAAAAKPWPATVRF-VHSRVEDLAGAGV
MMAR_0008|M.marinum_M LASELTMPGGEVVGTDRAAGAVSAASAAAAARGLSETVRFRLGDPADSSF
MUL_0010|M.ulcerans_Agy99 LASELTMPGGEVVGTDRAAGAVSAASATAAVRGLSETVRFRLGDPADLSF
Mb0862|M.bovis_AF2122/97 DLAARVVPG-SVTGVEPTDDALSLARAEAQLHRLSN-ISFTTSDVHKLDF
Rv0839|M.tuberculosis_H37Rv DLAARVVPG-SVTGVEPTDDALSLARAEAQLHRLSN-ISFTTSDVHKLDF
MAV_0854|M.avium_104 DLAARVAPG-QVTAVDQAADVLEVARAEAEQRNLSN-VSFGTADVHRLDF
Mvan_4620|M.vanbaalenii_PYR-1 RLRS---VGADIAAIDPSPPFIDAIRRSFPDVDVRQ------GPAEELPY
MSMEG_0098|M.smegmatis_MC2_155 QLARAAGLDGLALGVDISEPMLARAVAAEAGRQVGF----VRADAQQLPF
MLBr_00551|M.leprae_Br4923 PAAK---TGATIVSTDLTPELLQRSQARATELGLTL--EYREADAQALPF
MAB_4742c|M.abscessus_ATCC_199 REAGLNNLARLAQYLPLLSEVEQKILGCFRRGGGLQYQDYPRFHAVMGER
Mflv_1845|M.gilvum_PYR-GCK TG-PFDGILAAYLIRNVADRDGVLQELRALLRPGG----VFAAHEYSVR-
TH_3050|M.thermoresistible__bu GG-PFDAVFAAYLLRNLPDPDTQLTAFRDLLRPGG----ILAVHEYSVR-
MMAR_0008|M.marinum_M ER-PFDAIVGRYVLTFQADPAAMLHRLSQHLVPGG----IVAFHEPDWSA
MUL_0010|M.ulcerans_Agy99 ER-PFDAIVGRYVLTFQADLAAMLHRLSQRLVPGG----IVAFHEPDWSA
Mb0862|M.bovis_AF2122/97 PDDAFDVVHAHQVLQHVADPVRALQEMRRVCTPGG----IVAARDADYSG
Rv0839|M.tuberculosis_H37Rv PDDAFDVVHAHQVLQHVADPVRALQEMRRVCTPGG----IVAARDADYSG
MAV_0854|M.avium_104 ADDTFDVVHAHQVLQHLSDPVAALREMRRVCRPGG----IVAVRDADYAG
Mvan_4620|M.vanbaalenii_PYR-1 DSAAFDAALAQLVVHFMTDPVAGLRQMSRVTRPGG----VVAACVWDGPT
MSMEG_0098|M.smegmatis_MC2_155 RDEVFDAATSLAVFQLIPDPVAAVSEIVRVLKPGG----RVAIMVPTAG-
MLBr_00551|M.leprae_Br4923 GDGEFDVVMSAIGVMFAPDHRRAASELVRVCRPNG----TIGVISWTQEG
MAB_4742c|M.abscessus_ATCC_199 SREVFDKALIRTVLPLVEGLPEGLEGGLDVADFGCGSGYAINLMAQAFPA
** . . . .
Mflv_1845|M.gilvum_PYR-GCK -----DSRTATALWNLVSSAVIIPAGRLRSGD-----------------A
TH_3050|M.thermoresistible__bu -----DSAVATAVWNVVCGAVVIPLGRLRTGD-----------------A
MMAR_0008|M.marinum_M VRSSPPAPTHDRCCHWIREALDRTHTSWDMADRLHNAFTGAGLPAPTLTM
MUL_0010|M.ulcerans_Agy99 VRSSPPAPTHDRCCQWIREALDRAHTSWDMADRLHNAFTGAGLPAPTSTM
Mb0862|M.bovis_AF2122/97 FIWFPKLPALDRWLDLYERAARANGGEPDAGRRLLSWARAAGFDDVTPTA
Rv0839|M.tuberculosis_H37Rv FIWFPKLPALDRWLDLYERAARANGGEPDAGRRLLSWARAAGFDDVTPTA
MAV_0854|M.avium_104 FIWYPELPALDLWRDLYRRVARANRGEPDAGRRLLSWARQAGFDDITPTG
Mvan_4620|M.vanbaalenii_PYR-1 G---ALAPFWDAVHVVDPEAEDEALLSGARSGHLTELFEAAGLRDVVEDS
MSMEG_0098|M.smegmatis_MC2_155 ------------------------------------------------AV
MLBr_00551|M.leprae_Br4923 FFG--RMLATIRPYRPSLSAALPPAALWGREAYIRGLLDDTVTNVKTERR
MAB_4742c|M.abscessus_ATCC_199 SRFTGVDFSDEAIATAAADAERLGLRNASFLSADVATLNAPATYDLITAF
Mflv_1845|M.gilvum_PYR-GCK ELYRYLKRSVNEFDGAAEFRNRLQRNGFVDVTSHT---------MSGWQH
TH_3050|M.thermoresistible__bu ALYRYLRRSVNTFDGAERFRNRLRACGFTDVRSAT---------MPGWQR
MMAR_0008|M.marinum_M RTFIGAGRAAAVWLQALAD---LVETLLPTIQAQG---------VATATE
MUL_0010|M.ulcerans_Agy99 RTFIGAGRAAAVWLQALAD---LVETLLPTIQAQG---------VATATE
Mb0862|M.bovis_AF2122/97 SVWCFATASAREWWGLVWADRILQSDLAHQLVDSG---------LATAAQ
Rv0839|M.tuberculosis_H37Rv SVWCFATASAREWWGLVWADRILQSDLAHQLVDSG---------LATAAQ
MAV_0854|M.avium_104 SLWCYATPETRDWWGGMWADRILHSTVARDLVSLG---------LAAREQ
Mvan_4620|M.vanbaalenii_PYR-1 IAVDVVHPTFEQWWEPYTYGVGPAGDYVQQLDDEG---------RARLES
MSMEG_0098|M.smegmatis_MC2_155 KPVTFLARGGARIFGDDELGDIFENVGLVGVRVKT---------HGFIQW
MLBr_00551|M.leprae_Br4923 QLEVKRFNSAEGVHNYFKNHYGPTIEAYTNIGDNS---------VLAAEL
MAB_4742c|M.abscessus_ATCC_199 DTIHDQAHPAAVLANIHRALRPGGVLLMADIKASSRLEDNIGVAMSTYRY
:
Mflv_1845|M.gilvum_PYR-GCK DIVHTFLGRTPQ--------------------------------------
TH_3050|M.thermoresistible__bu DIVHTFLARRPG--------------------------------------
MMAR_0008|M.marinum_M VGIETLTQRLLEEVSDLGATVIGRSEVGAWTRTPRSG-------------
MUL_0010|M.ulcerans_Agy99 VGIETLTQRLLEEVSDLGATVIGRSEVGAWTRTPHSG-------------
Mb0862|M.bovis_AF2122/97 LEEISTAWREWAAAPD-GWLAIPHGEILCRA-------------------
Rv0839|M.tuberculosis_H37Rv LEEISTAWREWAAAPD-GWLAIPHGEILCRA-------------------
MAV_0854|M.avium_104 LEEISAAWREWAAAPD-GWIAIPHGEIICRA-------------------
Mvan_4620|M.vanbaalenii_PYR-1 AARERLGKGPFT-VTAVAWAARGTA-------------------------
MSMEG_0098|M.smegmatis_MC2_155 VRGTKL--------------------------------------------
MLBr_00551|M.leprae_Br4923 DAQLTELAQKYLTNSTMGWEYLLLTAQKH---------------------
MAB_4742c|M.abscessus_ATCC_199 TVSLTHCMPVSLGLEGAGLGTMWGRQLAESMLAEAGFSDVTVREIEQDTS
Mflv_1845|M.gilvum_PYR-GCK --------
TH_3050|M.thermoresistible__bu --------
MMAR_0008|M.marinum_M --------
MUL_0010|M.ulcerans_Agy99 --------
Mb0862|M.bovis_AF2122/97 --------
Rv0839|M.tuberculosis_H37Rv --------
MAV_0854|M.avium_104 --------
Mvan_4620|M.vanbaalenii_PYR-1 --------
MSMEG_0098|M.smegmatis_MC2_155 --------
MLBr_00551|M.leprae_Br4923 --------
MAB_4742c|M.abscessus_ATCC_199 NYFYIARR