For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VNDKRRAIYTHGYHESVLRSHRRRTAENSAGYLLPYLVPGLSVLDVGCGPGTITVDLAARVVPGSVTGVE PTDDALSLARAEAQLHRLSNISFTTSDVHKLDFPDDAFDVVHAHQVLQHVADPVRALQEMRRVCTPGGIV AARDADYSGFIWFPKLPALDRWLDLYERAARANGGEPDAGRRLLSWARAAGFDDVTPTASVWCFATASAR EWWGLVWADRILQSDLAHQLVDSGLATAAQLEEISTAWREWAAAPDGWLAIPHGEILCRA
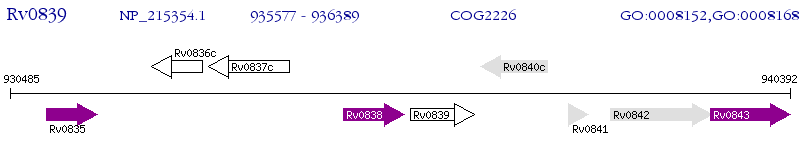
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0839 | - | - | 100% (270) | hypothetical protein Rv0839 |
| M. tuberculosis H37Rv | Rv1523 | - | 5e-10 | 35.35% (99) | methyltransferase |
| M. tuberculosis H37Rv | Rv2258c | - | 3e-09 | 31.43% (105) | transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0862 | - | 1e-160 | 100.00% (270) | hypothetical protein Mb0862 |
| M. gilvum PYR-GCK | Mflv_2768 | - | 1e-09 | 35.00% (100) | methyltransferase type 11 |
| M. leprae Br4923 | MLBr_00551 | - | 3e-08 | 31.25% (112) | putative methyltransferase |
| M. abscessus ATCC 19977 | MAB_4742c | - | 2e-11 | 35.85% (106) | hypothetical protein MAB_4742c |
| M. marinum M | MMAR_4799 | - | 1e-124 | 75.46% (269) | hypothetical protein MMAR_4799 |
| M. avium 104 | MAV_0854 | - | 1e-123 | 75.19% (270) | methyltransferase-UbiE family protein |
| M. smegmatis MC2 155 | MSMEG_0098 | - | 2e-13 | 42.31% (104) | methyltransferase |
| M. thermoresistible (build 8) | TH_2189 | - | 7e-08 | 28.57% (140) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0368 | - | 1e-124 | 75.84% (269) | hypothetical protein MUL_0368 |
| M. vanbaalenii PYR-1 | Mvan_4620 | - | 2e-11 | 28.50% (207) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0839|M.tuberculosis_H37Rv --------------------------------------------------
Mb0862|M.bovis_AF2122/97 --------------------------------------------------
MMAR_4799|M.marinum_M --------------------------------------------------
MUL_0368|M.ulcerans_Agy99 --------------------------------------------------
MAV_0854|M.avium_104 --------------------------------------------------
Mflv_2768|M.gilvum_PYR-GCK --------------------------------------------------
MAB_4742c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_4620|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0098|M.smegmatis_MC2_155 --------------------------------------------------
TH_2189|M.thermoresistible__bu VIRRLMATALTALTTAVLAGCGDAGSWVEAKPAEGWSAQYGDAANSSYTS
MLBr_00551|M.leprae_Br4923 --------------------------------------------------
Rv0839|M.tuberculosis_H37Rv --------------------------------------------------
Mb0862|M.bovis_AF2122/97 --------------------------------------------------
MMAR_4799|M.marinum_M --------------------------------------------------
MUL_0368|M.ulcerans_Agy99 --------------------------------------------------
MAV_0854|M.avium_104 --------------------------------------------------
Mflv_2768|M.gilvum_PYR-GCK --------------------------------------------------
MAB_4742c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_4620|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0098|M.smegmatis_MC2_155 --------------------------------------------------
TH_2189|M.thermoresistible__bu AAGAEALTLEWSRSVKGELAAQVAVGASGYLAVNAQTPAGCSLMVWEYAN
MLBr_00551|M.leprae_Br4923 --------------------------------------------------
Rv0839|M.tuberculosis_H37Rv --------------------------------------------------
Mb0862|M.bovis_AF2122/97 --------------------------------------------------
MMAR_4799|M.marinum_M --------------------------------------------------
MUL_0368|M.ulcerans_Agy99 --------------------------------------------------
MAV_0854|M.avium_104 --------------------------------------------------
Mflv_2768|M.gilvum_PYR-GCK --------------------------------------------------
MAB_4742c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_4620|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0098|M.smegmatis_MC2_155 --------------------------------------------------
TH_2189|M.thermoresistible__bu SARQRWCTRLVQGGGRTSPLLDGFDNVYIGQPGAILSFPPTQWIRWRKPV
MLBr_00551|M.leprae_Br4923 --------------------------------------------------
Rv0839|M.tuberculosis_H37Rv --------------------------------------------------
Mb0862|M.bovis_AF2122/97 --------------------------------------------------
MMAR_4799|M.marinum_M --------------------------------------------------
MUL_0368|M.ulcerans_Agy99 --------------------------------------------------
MAV_0854|M.avium_104 --------------------------------------------------
Mflv_2768|M.gilvum_PYR-GCK --------------------------------------------------
MAB_4742c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_4620|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0098|M.smegmatis_MC2_155 --------------------------------------------------
TH_2189|M.thermoresistible__bu IGMPTTPRILAPGELLVVTHLGQVLLFDAHRGTVTGTPLDLVAGVDPTDS
MLBr_00551|M.leprae_Br4923 --------------------------------------------------
Rv0839|M.tuberculosis_H37Rv --------------------------------------------------
Mb0862|M.bovis_AF2122/97 --------------------------------------------------
MMAR_4799|M.marinum_M --------------------------------------------------
MUL_0368|M.ulcerans_Agy99 --------------------------------------------------
MAV_0854|M.avium_104 --------------------------------------------------
Mflv_2768|M.gilvum_PYR-GCK ----------------------------MTTLAENSAPSSETTEEFAQRI
MAB_4742c|M.abscessus_ATCC_199 ----------------------------MTAPSANPVP--ETTEEFSERL
Mvan_4620|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0098|M.smegmatis_MC2_155 -------------------------MDTALRRAMDLLIDPPAEPDFSKGY
TH_2189|M.thermoresistible__bu ERGLADCAGARXXXCWPPPRPPSTVTPRARDAARHPTARLPAMTETTEPG
MLBr_00551|M.leprae_Br4923 ------------------------------------------MNLYTPIH
Rv0839|M.tuberculosis_H37Rv ----MNDKRRAIYTHGYHESVLR---------------------------
Mb0862|M.bovis_AF2122/97 ----MNDKRRAIYTHGYHESVLR---------------------------
MMAR_4799|M.marinum_M ----MNDERRAVYTHGHHESVLR---------------------------
MUL_0368|M.ulcerans_Agy99 ----MNDERRAVYTHGHHESVLR---------------------------
MAV_0854|M.avium_104 ----MNDQHRATYTHGHHESVLR---------------------------
Mflv_2768|M.gilvum_PYR-GCK VGALDAASLALLVSIGHQTHLFDTMA------------------------
MAB_4742c|M.abscessus_ATCC_199 TAAIDNAALVLLMSIGHQTGLLDTLA------------------------
Mvan_4620|M.vanbaalenii_PYR-1 --------MSFVVASEAYGRFMG---------------------------
MSMEG_0098|M.smegmatis_MC2_155 LDLLGRGDSTAPKNTGLIQKAWASPLGS--------------------KL
TH_2189|M.thermoresistible__bu IGSEGAGDLPAPNPHATAEQVEAAMRDSKLAQILYHDWEAESYDDKWSIS
MLBr_00551|M.leprae_Br4923 EDQSLAAIHRAMWALGDYALMAE---------------------------
Rv0839|M.tuberculosis_H37Rv ------SHRRRTAENSAGYLLPYLVPGLSVLDVG------------CGPG
Mb0862|M.bovis_AF2122/97 ------SHRRRTAENSAGYLLPYLVPGLSVLDVG------------CGPG
MMAR_4799|M.marinum_M ------GHRQRTVANSAGYLSPHLRAGLSVLDIG------------CGPG
MUL_0368|M.ulcerans_Agy99 ------GHRQRTVANSAGYLSPHLRAGLSVLDIG------------CGPG
MAV_0854|M.avium_104 ------SHRRRTAEDSAGYLLAHLTPGLSVLDVG------------CGPG
Mflv_2768|M.gilvum_PYR-GCK ---NLPPSTSTQIADAAGLNERYVREWLGGVAAGRIVDYDPAAQTFSLPA
MAB_4742c|M.abscessus_ATCC_199 ---ASAGSTSEQLADAAGLEERYVREWLAGMTVAHVVDYEPVSAIYTLPS
Mvan_4620|M.vanbaalenii_PYR-1 --------RYAEPLAEAFVAFAGVGVGDKVLDVG------------CGPG
MSMEG_0098|M.smegmatis_MC2_155 YDRAQLLNRRLVASTRPPVEWLRIPPGGTVLDIG------------CGPG
TH_2189|M.thermoresistible__bu YDQRCVDYARGRFDAIVPDHVQRELPYDNALELG------------CGTG
MLBr_00551|M.leprae_Br4923 --------EVMAPLGPILVTATGIRPGMQVLDVA------------AGSG
: . ..
Rv0839|M.tuberculosis_H37Rv TITVDLAARVVPGSVTG-VEPTDDALSLARAEAQLHR-----LSNISFTT
Mb0862|M.bovis_AF2122/97 TITVDLAARVVPGSVTG-VEPTDDALSLARAEAQLHR-----LSNISFTT
MMAR_4799|M.marinum_M TITVDLAARVAPATVTA-VEPTDAALNLARAEVQRCD-----VSNVAFVT
MUL_0368|M.ulcerans_Agy99 TITVDLAARVAPGTVTA-VEPTDAALNLGRAEAQRCD-----VSNVAFVT
MAV_0854|M.avium_104 TITADLAARVAPGQVTA-VDQAADVLEVARAEAEQRN-----LSNVSFGT
Mflv_2768|M.gilvum_PYR-GCK HRAAVLTRTAGPDNLAR-VTQFIPLLSEVEQKVIACFRDGGGLSYAEYPR
MAB_4742c|M.abscessus_ATCC_199 HRAATLTREAGLNNLAR-LAQYLPLLSEVEQKILGCFRRGGGLQYQDYPR
Mvan_4620|M.vanbaalenii_PYR-1 ALTARLRSVG--ADIAA-IDPSPPFIDAIRRS----------FPDVDVRQ
MSMEG_0098|M.smegmatis_MC2_155 NITAQLARAAGLDGLALGVDISEPMLARAVAAEAG--------RQVGFVR
TH_2189|M.thermoresistible__bu FFLLNLIQSGVARRGSV-TDLSPGMVRVATRNGRSLG------LDVDGRV
MLBr_00551|M.leprae_Br4923 NISLPAAKTG--ATIVS-TDLTPELLQRSQARATELG------LTLEYRE
:
Rv0839|M.tuberculosis_H37Rv SDVHKLDFPDDAFDVVHAHQVLQHVADPVRALQEMRRVCTPGG-------
Mb0862|M.bovis_AF2122/97 SDVHKLDFPDDAFDVVHAHQVLQHVADPVRALQEMRRVCTPGG-------
MMAR_4799|M.marinum_M SDVHALDFPDDVFDVVHAHQVLQHVADPVQALREMKRVCAPGG-------
MUL_0368|M.ulcerans_Agy99 SDVHALDFPDDVFDVVHAHQVLQHVADPVQALREMKRVGAPGG-------
MAV_0854|M.avium_104 ADVHRLDFADDTFDVVHAHQVLQHLSDPVAALREMRRVCRPGG-------
Mflv_2768|M.gilvum_PYR-GCK FHTLMAEQSGEVFDAALVDGILPMAEGLTDRLRQGVDVADFGCGSGHA-V
MAB_4742c|M.abscessus_ATCC_199 FHAVMGERSREVFDKALIRTVLPLVEGLPEGLEGGLDVADFGCGSGYA-I
Mvan_4620|M.vanbaalenii_PYR-1 GPAEELPYDSAAFDAALAQLVVHFMTDPVAGLRQMSRVTRPGG-------
MSMEG_0098|M.smegmatis_MC2_155 ADAQQLPFRDEVFDAATSLAVFQLIPDPVAAVSEIVRVLKPGG-------
TH_2189|M.thermoresistible__bu ADAEGIPYDDNTFDLVVGHAVLHHIPDVELSLREVVRVLRPGGRFVFAGE
MLBr_00551|M.leprae_Br4923 ADAQALPFGDGEFDVVMSAIGVMFAPDHRRAASELVRVCRPNG-------
. ** . . . * .
Rv0839|M.tuberculosis_H37Rv ---IVAARDADYSGFIWFPK--LPALDRWLDLYERAARANGGEPDAGRRL
Mb0862|M.bovis_AF2122/97 ---IVAARDADYSGFIWFPK--LPALDRWLDLYERAARANGGEPDAGRRL
MMAR_4799|M.marinum_M ---LVAARDADYAGFIWYPQ--LPALDHWLKLYQRAARANGGEPDAGRRL
MUL_0368|M.ulcerans_Agy99 ---LVAARDADYAGFIWYPQ--LPALDHWLQLYQRAARANGGEPDAGRRL
MAV_0854|M.avium_104 ---IVAVRDADYAGFIWYPE--LPALDLWRDLYRRVARANRGEPDAGRRL
Mflv_2768|M.gilvum_PYR-GCK NVMAKAFPASRFTGIDFSEEGLAAGRDEARRLGLTNASFNATDVATYDAA
MAB_4742c|M.abscessus_ATCC_199 NLMAQAFPASRFTGVDFSDEAIATAAADAERLGLRNASFLSADVATLNAP
Mvan_4620|M.vanbaalenii_PYR-1 ----------VVAACVWDGP-TGALAPFWDAVHVVDPEAEDEALLSGARS
MSMEG_0098|M.smegmatis_MC2_155 ------------------------------RVAIMVPTAGAVKPVTFLAR
TH_2189|M.thermoresistible__bu PTTVGNSYARALSTLTWHVATTVTRLPGLQGWRRPQAELDESSRAAALEA
MLBr_00551|M.leprae_Br4923 ----------TIGVISWTQEGFFGRMLATIRPYRPSLSAALPPAALWGRE
Rv0839|M.tuberculosis_H37Rv LSWARAAGFDDVTPTAS---VWCFATASAREWWGLVWADRILQSDLAHQL
Mb0862|M.bovis_AF2122/97 LSWARAAGFDDVTPTAS---VWCFATASAREWWGLVWADRILQSDLAHQL
MMAR_4799|M.marinum_M LSWARQAGFADITPTGS---MWCYATDEMRAWWGGMWAERILRSDLSAQL
MUL_0368|M.ulcerans_Agy99 LSWARQAGFADITPTGS---MWCYATDEMRAWWGGMWADRILRSDLSAQL
MAV_0854|M.avium_104 LSWARQAGFDDITPTGS---LWCYATPETRDWWGGMWADRILHSTVARDL
Mflv_2768|M.gilvum_PYR-GCK ESFDVITAFDAIHDQAHPARVLANIHAALRPGGTFLMVDIKSSSRLENNI
MAB_4742c|M.abscessus_ATCC_199 ATYDLITAFDTIHDQAHPAAVLANIHRALRPGGVLLMADIKASSRLEDNI
Mvan_4620|M.vanbaalenii_PYR-1 GHLTELFEAAGLRDVVEDSIAVDVVHPTFEQWWEPYTYGVGPAGDYVQQL
MSMEG_0098|M.smegmatis_MC2_155 G-GARIFGDDELGDIFEN--------------------------------
TH_2189|M.thermoresistible__bu VVDLHTFEPSDLERMARNAGAVEVATASEEFTAAMLGWPVRTFEAAVPPG
MLBr_00551|M.leprae_Br4923 AYIRGLLDDTVTNVKTERRQLEVKRFNSAEGVHNYFKNHYGPTIEAYTNI
Rv0839|M.tuberculosis_H37Rv ------------------VDSGLATAAQLEEISTAWREWAAAPDGWLAIP
Mb0862|M.bovis_AF2122/97 ------------------VDSGLATAAQLEEISTAWREWAAAPDGWLAIP
MMAR_4799|M.marinum_M ------------------LGSGMASAADLEAISQAWRDWAAAPDGWLGIP
MUL_0368|M.ulcerans_Agy99 ------------------LGSGMASAADLEAISQAWRDWAAAPDGWLGIP
MAV_0854|M.avium_104 ------------------VSLGLAAREQLEEISAAWREWAAAPDGWIAIP
Mflv_2768|M.gilvum_PYR-GCK GVPFAPYLYTVSAMHCMSVSLGLDGDGLGTCWGREVATTMLADAGFGDVQ
MAB_4742c|M.abscessus_ATCC_199 GVAMSTYRYTVSLTHCMPVSLGLEGAGLGTMWGRQLAESMLAEAGFSDVT
Mvan_4620|M.vanbaalenii_PYR-1 D------------------DEGRARLESAARERLGKGPFTVTAVAWAARG
MSMEG_0098|M.smegmatis_MC2_155 --------------------VGLVG-------------VRVKTHGFIQWV
TH_2189|M.thermoresistible__bu KLG-------------WNWAKFAFGSWTALSWVDANVWRRVMPKGWFYNV
MLBr_00551|M.leprae_Br4923 G-----------------DNSVLAAELDAQLTELAQKYLTNSTMGWEYLL
.:
Rv0839|M.tuberculosis_H37Rv HGEILCRA---------
Mb0862|M.bovis_AF2122/97 HGEILCRA---------
MMAR_4799|M.marinum_M NGEILCRV---------
MUL_0368|M.ulcerans_Agy99 NGEILCRV---------
MAV_0854|M.avium_104 HGEIICRA---------
Mflv_2768|M.gilvum_PYR-GCK VREIESDPINYYYIARK
MAB_4742c|M.abscessus_ATCC_199 VREIEQDTSNYFYIARR
Mvan_4620|M.vanbaalenii_PYR-1 TA---------------
MSMEG_0098|M.smegmatis_MC2_155 RGTKL------------
TH_2189|M.thermoresistible__bu MVTGVKPS---------
MLBr_00551|M.leprae_Br4923 LTAQKH-----------