For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGRVQDKVVLVTGGARGQGRSHAVKLAEEGADIILFDICHDIETNEYPLATSRDLEEAGLEVEKTGRKAY TAEVDVRDRAAVSRELANAVAEFGKLDVVVANAGICPLGAHLPVQAFADAFDVDFVGVINTVHAALPYLT SGASIITTGSVAGLIAAAQPPGAGGPQGPGGAGYSYAKQLVDSYTLQLAAQLAPQSIRANVIHPTNVNTD MLNSAPMYRQFRPDLEAPSRADALLAFPAMQAMPTPYVEASDISNAVCFLASDESRYVTGLQFKVDAGAM LKF
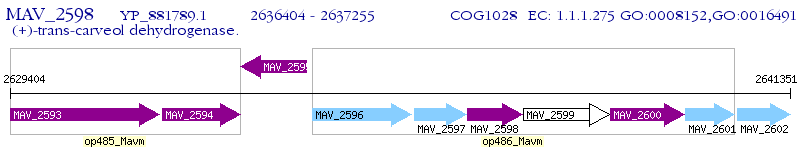
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2598 | - | - | 100% (283) | carveol dehydrogenase |
| M. avium 104 | MAV_1881 | - | e-129 | 81.05% (285) | carveol dehydrogenase ((+)-trans-carveol dehydrogenase) |
| M. avium 104 | MAV_2946 | - | 3e-92 | 61.84% (283) | carveol dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2771 | - | 2e-48 | 41.20% (284) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_2500 | - | 2e-84 | 56.89% (283) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2750 | - | 2e-48 | 41.20% (284) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_01094 | - | 4e-15 | 29.17% (216) | short chain alcohol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0072c | - | 1e-50 | 41.84% (282) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_4754 | - | 6e-55 | 44.44% (288) | short-chain type dehydrogenase/reductase |
| M. smegmatis MC2 155 | MSMEG_4801 | - | 7e-85 | 56.38% (282) | carveol dehydrogenase |
| M. thermoresistible (build 8) | TH_3929 | - | 1e-101 | 64.54% (282) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3392 | fabG | 2e-44 | 36.62% (284) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. vanbaalenii PYR-1 | Mvan_2006 | - | 1e-85 | 55.32% (282) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_2598|M.avium_104 ----MG----RVQDKVVLVTGGARGQGRSHAVKLAEEGADIILFDICHD-
TH_3929|M.thermoresistible__bu ----MN----RLTGKVVAITGAARGQGRSHAVHMADEGADIIALDICAD-
Mvan_2006|M.vanbaalenii_PYR-1 ----MG----KLDNKVAVVSGAARGQGRSHAVSLAAEGAHIIALDICAD-
Mflv_2500|M.gilvum_PYR-GCK ----MG----RVQGKVAFITGAARGQGRSHAVRLAEEGADIIAVDICRD-
MSMEG_4801|M.smegmatis_MC2_155 ----MGSAEGRVAGKVAFITGAARGQGRSHAVRLAEEGADIIAVDLCQD-
MAB_0072c|M.abscessus_ATCC_199 ----MTG---RLTGKVALVTGAARGIGRAQAVRFAQEGADIIALDICAP-
MMAR_4754|M.marinum_M MTEQLGG---RVAGKVAFITGAARGQGRSHAVRLAQEGADIIAVDVCKPI
Mb2771|M.bovis_AF2122/97 ---MIDR---PLEGKVAFITGAARGLGRAHAVRLAADGANIIAVDICEQ-
Rv2750|M.tuberculosis_H37Rv ---MIDR---PLEGKVAFITGAARGLGRAHAVRLAADGANIIAVDICEQ-
MUL_3392|M.ulcerans_Agy99 MSRPLDG---PLSGRVAFITGAARGQGRAHALRLARDGADVIAVDLCDQ-
MLBr_01094|M.leprae_Br4923 MEG--------FAGKVAVVTGAGSGIGQALAIELARSGAKLAISDVDGE-
. .:*. ::*.. * *:: *: :* .**.: *:
MAV_2598|M.avium_104 IETNEYPLATSRDLEEAGLEVEKTGRKAYTAEVDVRDRAAVSRELANAVA
TH_3929|M.thermoresistible__bu IATNEYPLATREDLEETARLVEKAGRRAITAEVDVRDRARLKQVLDDAVA
Mvan_2006|M.vanbaalenii_PYR-1 LEGNTYPLSRPEDLDETARLAEKEGVRVHTAIVDVRERAAVWKAVADGVD
Mflv_2500|M.gilvum_PYR-GCK YDTVGYRMATPEDLEETKNFVEKTGQRIVTAQADVRNEAELRAALEAGLA
MSMEG_4801|M.smegmatis_MC2_155 IDSIGYSLATPEDLEETARYVEKAGRRIVTAQADVRDAAQLKEALERGVS
MAB_0072c|M.abscessus_ATCC_199 VHTTITPPATRADLDKTSELVESAGRRVIPGVVDVRNLRDVQTFTQDAVT
MMAR_4754|M.marinum_M VENTTIPASTPEDLAETADLVKGHNRRIFTAEADVRDYDALKAAVDAGVD
Mb2771|M.bovis_AF2122/97 IASVPYPLSTADDLAATVELVEDAGGGIVARQGDVRDRASLSVALQAGLD
Rv2750|M.tuberculosis_H37Rv IASVPYPLSTADDLAATVELVEDAGGGIVARQGDVRDRASLSVALQAGLD
MUL_3392|M.ulcerans_Agy99 IASVPYPLGTAEELATTVKLVEDTGARIVASQADVRDREALAAALQAGID
MLBr_01094|M.leprae_Br4923 ------------GLAQTEGQLKAIGASARTDRLDVTEREAFLTYADVVHE
* : : . . ** : .
MAV_2598|M.avium_104 EFGKLDVVVANAGICP---LGAHLPVQAFADAFDVDFVGVINTVHAALPY
TH_3929|M.thermoresistible__bu ELGGLHVVVANAGICP---QGNHLPYKAYIDAFDVDFVGVVNTIHVGLDY
Mvan_2006|M.vanbaalenii_PYR-1 ALGRLDIVVANAGICP---MGRGQTIQSWSDAIDTDLVGVFNVIQASLPH
Mflv_2500|M.gilvum_PYR-GCK EFGKVDIVVAQAGIAA---MKGQPAMQAWTDGINTNFVGTINAIQVALPH
MSMEG_4801|M.smegmatis_MC2_155 ELGKLDIVVAQAGIAA---MKGNPPLQAWTDGINTNLVGTINAIQVALPH
MAB_0072c|M.abscessus_ATCC_199 EFGGLDIVCATAGITSRRMLLDMP-ESDWQTMLDVNLTGVWHTTRAATPH
MMAR_4754|M.marinum_M ELGRLDIIVANAGIGNGGATLDKTSEHDWQEMIDVNLSGVWKSVKAAVPH
Mb2771|M.bovis_AF2122/97 EFGRLDIVVANAGIA-----MMQAGDDGWRDVIDVNLTGVFHTVQVAIPT
Rv2750|M.tuberculosis_H37Rv EFGRLDIVVANAGIA-----MMQAGDDGWRDVIDVNLTGVFHTVQVAIPT
MUL_3392|M.ulcerans_Agy99 ELGQVDIVVANAGIA-----PMQSGDDGWRDVIDVNLSGAYYTVEVAIPT
MLBr_01094|M.leprae_Br4923 NFGKVNQIYNNAGIAFTG-DVEVSHFKDIERVMDVDYWGVVNGTKAFLPY
:* :. : *** ::.: *. ..
MAV_2598|M.avium_104 LTS---GASIITTGSVAGLIAAAQPPGAGGPQGPGGAGYSYAKQLVDSYT
TH_3929|M.thermoresistible__bu LGE---GGSVIVTGSIAGLVDQKDMANGGGPSGPGGAGYGMAKKMVRDYT
Mvan_2006|M.vanbaalenii_PYR-1 VND---GASIIATGSLAAQLGSATN------QGPGGSAYSLAKQVVAHYV
Mflv_2500|M.gilvum_PYR-GCK LKE---GASIIATASAAALMDAHNKPNPG--GDPGGMGYMISKRLISEYV
MSMEG_4801|M.smegmatis_MC2_155 LSE---GASIVATASAAALMDAHNKPNPG--ADPGGMGYMVSKRMISEYV
MAB_0072c|M.abscessus_ATCC_199 LIARG-GGAMVLVSSIAGLRG-------LVGVS----HYVAAKHGVVGLM
MMAR_4754|M.marinum_M LIAGGNGGSIVLTSSVGGMKA-------YPHCG----NYVAAKHGVVGLM
Mb2771|M.bovis_AF2122/97 LIEQGTGGSIVLISSAAGLVG-------IGSSDPGSLGYAAAKHGVVGLM
Rv2750|M.tuberculosis_H37Rv LIEQGTGGSIVLISSAAGLVG-------IGSSDPGSLGYAAAKHGVVGLM
MUL_3392|M.ulcerans_Agy99 MIEQGRGGSIVLISSAAGLVG-------ISSADAGAIGYVASKHALVGLM
MLBr_01094|M.leprae_Br4923 LISSG-DGHVINISSVFGLFS-----------VPGQAAYNSAKFAVRGFT
: .. :: .* . * :* :
MAV_2598|M.avium_104 LQLAAQLAPQS--IRANVIHPTNVNTDMLNSAPMYRQFRPDLEAPSRADA
TH_3929|M.thermoresistible__bu KALALTLAPHK--IRINAVHPTNVNTDMLHNMSMYRVFRPDLTEPTREDA
Mvan_2006|M.vanbaalenii_PYR-1 NDLSIQLAKRM--IRVNAIHPTNVNTDMLHNEGLYRVFRPDLKEPTREEA
Mflv_2500|M.gilvum_PYR-GCK HALATELAVRG--IRANVIHPTNCNTDMLQSEPMYRSFRPDLENPTRADA
MSMEG_4801|M.smegmatis_MC2_155 HYLATELTPLG--IRANVIHPTNCNTDMLQSEPMYRSFRPDLEHPTRADA
MAB_0072c|M.abscessus_ATCC_199 RSLAIELAPHK--IRVNTVHPTNVDTPMIQNEFVRTLFRPDLEQPTREDF
MMAR_4754|M.marinum_M RSFAVELGQHM--IRVNSVHPTHVRTPMLHNEGTFKMFRPDLENPGPDDM
Mb2771|M.bovis_AF2122/97 RAYANHLAPQN--IRVNSVHPCGVDTPMINNEFFQQWLT-------TADM
Rv2750|M.tuberculosis_H37Rv RAYANHLAPQN--IRVNSVHPCGVDTPMINNEFFQQWLT-------TADM
MUL_3392|M.ulcerans_Agy99 RVYANLLAPHS--IRVNSLHPSGVDPPMINNEFIRHWLAD-----LVAET
MLBr_01094|M.leprae_Br4923 EALREEMALAGRPVNVTTVYPGGIKTAIARNATAAEGLDVS-KIASRFDT
: :. . ::* . : .. : :
MAV_2598|M.avium_104 LLAFPAMQAMPTPYVEASDISNAVCFLASDESRYVTGLQFKVDAGAMLKF
TH_3929|M.thermoresistible__bu EVVFPLLQAMPIPYVEPDDVSHAVVYLASDESRYVTGQQLFVDAGAGLKM
Mvan_2006|M.vanbaalenii_PYR-1 EEAFPAMQAMPIPYIEPRDVSNAVVFLAGDDSRYITGTQLRIDAGGYVKA
Mflv_2500|M.gilvum_PYR-GCK EPVFGVQQAMKVNFIEPEDISNAVLWLASDESRYVTGMQLRVDAGGYLKW
MSMEG_4801|M.smegmatis_MC2_155 EPVFYVQQAMKVPWVEPSDISNAVLWLASDESRFVTGMQLRVDAGGYLKW
MAB_0072c|M.abscessus_ATCC_199 AAAAKTMNLLDLPWVEPLDIANASLFLASDEARYITSVSLPVDAGSTQR-
MMAR_4754|M.marinum_M APICQLFHTLPIPWVEAEDISNAVLFLASDESRYITGVTLPVDAGGCLK-
Mb2771|M.bovis_AF2122/97 DAPHNLGNALPVELVQPTDIANAVAWLASEEARYVTGVTLPVDAGFVNKR
Rv2750|M.tuberculosis_H37Rv DAPHNLGNALPVELVQPTDIANAVAWLASEEARYVTGVTLPVDAGFVNKR
MUL_3392|M.ulcerans_Agy99 GSGPGAGNALPVQILQADDIAGALAWLVSDEARYITGVALPVDAGCVNKR
MLBr_01094|M.leprae_Br4923 WVAHTSPQHAARIILKAVRKKKARVLVGPDAKVANVVVRFSGGAGYQRLF
: ::. * : : . : .**
MAV_2598|M.avium_104 ------------
TH_3929|M.thermoresistible__bu GV----------
Mvan_2006|M.vanbaalenii_PYR-1 VPWKG-------
Mflv_2500|M.gilvum_PYR-GCK YDYHV-------
MSMEG_4801|M.smegmatis_MC2_155 YDYHV-------
MAB_0072c|M.abscessus_ATCC_199 ------------
MMAR_4754|M.marinum_M ------------
Mb2771|M.bovis_AF2122/97 ------------
Rv2750|M.tuberculosis_H37Rv ------------
MUL_3392|M.ulcerans_Agy99 ------------
MLBr_01094|M.leprae_Br4923 AQVASRLILNQR