For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TGRLTGKVALVTGAARGIGRAQAVRFAQEGADIIALDICAPVHTTITPPATRADLDKTSELVESAGRRVI PGVVDVRNLRDVQTFTQDAVTEFGGLDIVCATAGITSRRMLLDMPESDWQTMLDVNLTGVWHTTRAATPH LIARGGGAMVLVSSIAGLRGLVGVSHYVAAKHGVVGLMRSLAIELAPHKIRVNTVHPTNVDTPMIQNEFV RTLFRPDLEQPTREDFAAAAKTMNLLDLPWVEPLDIANASLFLASDEARYITSVSLPVDAGSTQR*
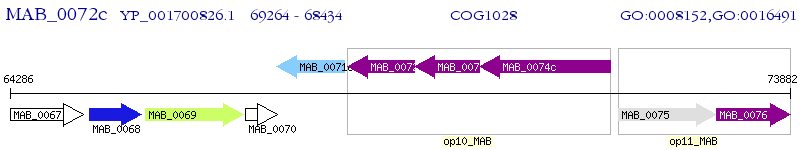
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0072c | - | - | 100% (276) | putative short chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_0073c | - | 2e-61 | 47.45% (274) | putative short chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_3081 | - | 7e-61 | 47.31% (279) | Short-chain dehydrogenase/reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2771 | - | 2e-62 | 50.18% (273) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_1589 | - | 3e-80 | 55.11% (274) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2750 | - | 2e-62 | 50.18% (273) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 5e-24 | 30.53% (262) | 3-oxoacyl- |
| M. marinum M | MMAR_4754 | - | 1e-77 | 54.95% (273) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_3017 | - | 1e-108 | 70.79% (267) | carveol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_4849 | - | 2e-79 | 53.96% (278) | carveol dehydrogenase |
| M. thermoresistible (build 8) | TH_3237 | - | 5e-80 | 55.27% (275) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3392 | fabG | 4e-56 | 44.44% (279) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. vanbaalenii PYR-1 | Mvan_2896 | - | 1e-105 | 68.00% (275) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2771|M.bovis_AF2122/97 ---MIDRPLEGKVAFITGAARGLGRAHAVR---LAADGANIIAVDICEQI
Rv2750|M.tuberculosis_H37Rv ---MIDRPLEGKVAFITGAARGLGRAHAVR---LAADGANIIAVDICEQI
MUL_3392|M.ulcerans_Agy99 MSRPLDGPLSGRVAFITGAARGQGRAHALR---LARDGADVIAVDLCDQI
MAV_3017|M.avium_104 --------------MITGAARGIGRAQAVR---FAQEGADIVALDLCGPV
Mvan_2896|M.vanbaalenii_PYR-1 -----MGRLSGKVAVITGAARGIGRAQAVR---FAQEGAGVIGIDLCGGV
MAB_0072c|M.abscessus_ATCC_199 ----MTGRLTGKVALVTGAARGIGRAQAVR---FAQEGADIIALDICAPV
Mflv_1589|M.gilvum_PYR-GCK ----MGGRVEGKVAFITGAARGQGRSHAVR---LAEEGADIIAIDVCGPI
TH_3237|M.thermoresistible__bu ----MAGRLEGKVAFITGAARGQGRAHAVR---MAQEGADIIAVDICRQI
MMAR_4754|M.marinum_M MTEQLGGRVAGKVAFITGAARGQGRSHAVR---LAQEGADIIAVDVCKPI
MSMEG_4849|M.smegmatis_MC2_155 ----MSGRVEGKVAFVTGAARGQGRSHALR---LAQEGADIIAIDICGPI
MLBr_01807|M.leprae_Br4923 ---------MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRL
..** * *:. : :. * :.: :. :
Mb2771|M.bovis_AF2122/97 AS---VPYPLSTADDLAATVELVEDAGGGIVARQGDVRDRASLSVALQAG
Rv2750|M.tuberculosis_H37Rv AS---VPYPLSTADDLAATVELVEDAGGGIVARQGDVRDRASLSVALQAG
MUL_3392|M.ulcerans_Agy99 AS---VPYPLGTAEELATTVKLVEDTGARIVASQADVRDREALAAALQAG
MAV_3017|M.avium_104 DT---VMVPPSTPDDLDHTASLVGEVGGRMHAELVDVRDLDGVQAATERG
Mvan_2896|M.vanbaalenii_PYR-1 DT---VVIPPSTPDDLAETARRVRAVGADVVTAIADVRDVDALGAAVDDG
MAB_0072c|M.abscessus_ATCC_199 HT---TITPPATRADLDKTSELVESAGRRVIPGVVDVRNLRDVQTFTQDA
Mflv_1589|M.gilvum_PYR-GCK STH--TDIPPATPDDLAQTVDLVKGTGRRIVSEQVDVRDFAAVKAAVDSG
TH_3237|M.thermoresistible__bu DS---VQIPLASPEDLAETADLVKGLNRRIYTAEVDVRDFDALKAAVDTG
MMAR_4754|M.marinum_M VEN--TTIPASTPEDLAETADLVKGHNRRIFTAEADVRDYDALKAAVDAG
MSMEG_4849|M.smegmatis_MC2_155 RPGAETAIPASTSDDLAETANLVKGLDRRVVTAEVDVRDAAAIKAAVDSG
MLBr_01807|M.leprae_Br4923 VAD----------GHRVAVTHRASVVPDWFFGVECDVTDNDAIGAAFKAV
. . . . ** : : . .
Mb2771|M.bovis_AF2122/97 LDEFGRLDIVVANAGIAMMQ-----AGDDGWRDVIDVNLTGVFHTVQVAI
Rv2750|M.tuberculosis_H37Rv LDEFGRLDIVVANAGIAMMQ-----AGDDGWRDVIDVNLTGVFHTVQVAI
MUL_3392|M.ulcerans_Agy99 IDELGQVDIVVANAGIAPMQ-----SGDDGWRDVIDVNLSGAYYTVEVAI
MAV_3017|M.avium_104 ARRFGGLDVVCATAGITSRAMTVEMN-ESVWRTMLDVNLTGVWHTCRAAA
Mvan_2896|M.vanbaalenii_PYR-1 VRALGGLDIVCATAGITSRGMAVEMS-EQTWQTMLDVNLTGVFHTCQVTA
MAB_0072c|M.abscessus_ATCC_199 VTEFGGLDIVCATAGITSRRMLLDMP-ESDWQTMLDVNLTGVWHTTRAAT
Mflv_1589|M.gilvum_PYR-GCK VEQLGRLDIVVANAGIGNGGQTLDHTSEEDWNDMIDVNLSGVWKSVKAAV
TH_3237|M.thermoresistible__bu VEQLGRLDIIVANAGIGNGGETLDKTSEGDWTDMIDVNLAGVWKTVKAGV
MMAR_4754|M.marinum_M VDELGRLDIIVANAGIGNGGATLDKTSEHDWQEMIDVNLSGVWKSVKAAV
MSMEG_4849|M.smegmatis_MC2_155 VEQLGRLDIIVANAGIGNGGDTLDQTSEEDWDEMIDINLSGVWKSVKAGV
MLBr_01807|M.leprae_Br4923 EEHQGPVEVLVANAGITSDMLIMRMS-EEQFQKVIDVNLTGVFRVAKLAS
* :::: *.*** : : ::*:**:*.: .
Mb2771|M.bovis_AF2122/97 PTLIEQGTGGSIVLISSAAGLVGIGSSDPGSLGYAAAKHGVVGLMRAYAN
Rv2750|M.tuberculosis_H37Rv PTLIEQGTGGSIVLISSAAGLVGIGSSDPGSLGYAAAKHGVVGLMRAYAN
MUL_3392|M.ulcerans_Agy99 PTMIEQGRGGSIVLISSAAGLVGISSADAGAIGYVASKHALVGLMRVYAN
MAV_3017|M.avium_104 PHLIARG-AGSMILTNSIAGLRGLVGVA----HYTAAKHGVVGLMQSLAH
Mvan_2896|M.vanbaalenii_PYR-1 PHLITRG-GGSIVLVSSIAGLRGLVGVA----HYTAAKHGVVGLMRSLAN
MAB_0072c|M.abscessus_ATCC_199 PHLIARG-GGAMVLVSSIAGLRGLVGVS----HYVAAKHGVVGLMRSLAI
Mflv_1589|M.gilvum_PYR-GCK PHLLAGGNGGSIILTSSVGGLKPYPHTG----HYIAAKHGVIGLMRTFAV
TH_3237|M.thermoresistible__bu PHILAGGRGGSIILTSSVGGLKAYPHTG----HYVAAKHGVVGLMRTFAV
MMAR_4754|M.marinum_M PHLIAGGNGGSIVLTSSVGGMKAYPHCG----NYVAAKHGVVGLMRSFAV
MSMEG_4849|M.smegmatis_MC2_155 PHIVAGGRGGSIVLTSSVGGLKAYPHCG----NYVAAKHGVVGLMRGFAV
MLBr_01807|M.leprae_Br4923 RNMIKQR-FGRYIFMGSVVGLSGVPGQA----NYAASKSGLIGMARSLAR
:: * :: .* *: * *:* .::*: : *
Mb2771|M.bovis_AF2122/97 HLAPQNIRVNSVHPCGVDTPMINNEFFQQWLT--------TADMDAPHNL
Rv2750|M.tuberculosis_H37Rv HLAPQNIRVNSVHPCGVDTPMINNEFFQQWLT--------TADMDAPHNL
MUL_3392|M.ulcerans_Agy99 LLAPHSIRVNSLHPSGVDPPMINNEFIRHWLAD------LVAETGSGPGA
MAV_3017|M.avium_104 ELAPHRVRVNCVHPTNVDTPLIQNDTVRSAFRPDLDRPPTRAEFAEAARA
Mvan_2896|M.vanbaalenii_PYR-1 ELARHRIRVNTVHPTNVDTPLIQNDAVRSAFRPDLDRPPTREEFAAAARP
MAB_0072c|M.abscessus_ATCC_199 ELAPHKIRVNTVHPTNVDTPMIQNEFVRTLFRPDLEQP-TREDFAAAAKT
Mflv_1589|M.gilvum_PYR-GCK ELGQHSIRVNAVCPTNVNTPLFMNEGTMKLFRPDLENP-GPDDLAVAAQF
TH_3237|M.thermoresistible__bu ELGQHNIRVNSVHPTNVNTPLFMNEPTMKLFRPDLENP-GPEDLKVVAQM
MMAR_4754|M.marinum_M ELGQHMIRVNSVHPTHVRTPMLHNEGTFKMFRPDLENP-GPDDMAPICQL
MSMEG_4849|M.smegmatis_MC2_155 ELGQHNIRVNTVHPTHVATPMLHNEGTFKMFRPDLENP-GPDDMAPICQM
MLBr_01807|M.leprae_Br4923 ELGSRSITANVVAPGFIDTEMTR-----------------AMDSKYQEAA
*. : : .* : * : . : :
Mb2771|M.bovis_AF2122/97 GNALPVELVQP-TDIANAVAWLASEEARYVTGVTLPVDAGFVNKR--
Rv2750|M.tuberculosis_H37Rv GNALPVELVQP-TDIANAVAWLASEEARYVTGVTLPVDAGFVNKR--
MUL_3392|M.ulcerans_Agy99 GNALPVQILQA-DDIAGALAWLVSDEARYITGVALPVDAGCVNKR--
MAV_3017|M.avium_104 MNLLQVPWVDP-VDVANAALFLASDEARYITAVTLPVDAGATQR---
Mvan_2896|M.vanbaalenii_PYR-1 MNMLETAWVDP-FDVANASLFLASDEARYITAAALPVDAGSTQRQPL
MAB_0072c|M.abscessus_ATCC_199 MNLLDLPWVEP-LDIANASLFLASDEARYITSVSLPVDAGSTQR---
Mflv_1589|M.gilvum_PYR-GCK MHVLPVGWVEP-RDISNAVLFLASDEARYVTGLPVTVDAGSMLK---
TH_3237|M.thermoresistible__bu MHTLPIGWVEP-EDIANAVLFLASDEARYITGVTLPVDGGSCLK---
MMAR_4754|M.marinum_M FHTLPIPWVEA-EDISNAVLFLASDESRYITGVTLPVDAGGCLK---
MSMEG_4849|M.smegmatis_MC2_155 FHTLPIPWVEP-IDISNAVLFLASDEARYITGVTLPVDAGSCLK---
MLBr_01807|M.leprae_Br4923 LQAIPLGRIGAVEEIAGAVSFLASEDAGYISGAVIPVDGGLGMGH--
: : : . :::.* :*.*::: *::. :.**.*