For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGKLDNKVAVVSGAARGQGRSHAVSLAAEGAHIIALDICADLEGNTYPLSRPEDLDETARLAEKEGVRVH TAIVDVRERAAVWKAVADGVDALGRLDIVVANAGICPMGRGQTIQSWSDAIDTDLVGVFNVIQASLPHVN DGASIIATGSLAAQLGSATNQGPGGSAYSLAKQVVAHYVNDLSIQLAKRMIRVNAIHPTNVNTDMLHNEG LYRVFRPDLKEPTREEAEEAFPAMQAMPIPYIEPRDVSNAVVFLAGDDSRYITGTQLRIDAGGYVKAVPW KG
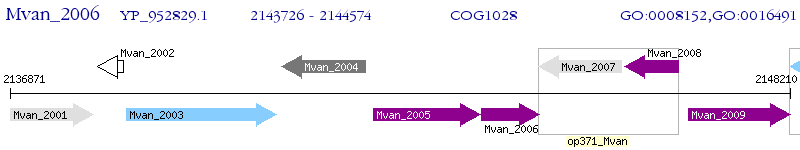
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2006 | - | - | 100% (282) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_4158 | - | 3e-81 | 52.50% (280) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_4222 | - | 1e-60 | 44.01% (284) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2771 | - | 4e-50 | 40.44% (272) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_4148 | - | 1e-84 | 55.48% (283) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2750 | - | 4e-50 | 40.44% (272) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_02565 | fabG | 1e-12 | 28.00% (275) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. abscessus ATCC 19977 | MAB_0072c | - | 1e-53 | 42.91% (275) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_4754 | - | 7e-63 | 46.45% (282) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_2946 | - | 2e-98 | 60.55% (289) | carveol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_4801 | - | 7e-82 | 53.76% (279) | carveol dehydrogenase |
| M. thermoresistible (build 8) | TH_3929 | - | 1e-86 | 56.38% (282) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3392 | fabG | 2e-45 | 37.23% (274) | 3-ketoacyl-(acyl-carrier-protein) reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2771|M.bovis_AF2122/97 --------------------------------------------------
Rv2750|M.tuberculosis_H37Rv --------------------------------------------------
MUL_3392|M.ulcerans_Agy99 --------------------------------------------------
Mvan_2006|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2946|M.avium_104 --------------------------------------------------
TH_3929|M.thermoresistible__bu --------------------------------------------------
MSMEG_4801|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_4148|M.gilvum_PYR-GCK --------------------------------------------------
MAB_0072c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_4754|M.marinum_M --------------------------------------------------
MLBr_02565|M.leprae_Br4923 MAPKCSSDLFSQIVSSGPGSFVAKQLGVPRPETLRRYRPSDAPLCGSLLI
Mb2771|M.bovis_AF2122/97 ------------------------------------MIDR-------PLE
Rv2750|M.tuberculosis_H37Rv ------------------------------------MIDR-------PLE
MUL_3392|M.ulcerans_Agy99 ---------------------------------MSRPLDG-------PLS
Mvan_2006|M.vanbaalenii_PYR-1 -----------------------------------------MG----KLD
MAV_2946|M.avium_104 -----------------------------------------MG----RVT
TH_3929|M.thermoresistible__bu -----------------------------------------MN----RLT
MSMEG_4801|M.smegmatis_MC2_155 -----------------------------------------MGSAEGRVA
Mflv_4148|M.gilvum_PYR-GCK -----------------------------------------MTG---KLE
MAB_0072c|M.abscessus_ATCC_199 -----------------------------------------MTG---RLT
MMAR_4754|M.marinum_M -------------------------------------MTEQLGG---RVA
MLBr_02565|M.leprae_Br4923 GGDGRLVEPLRATLEKDYDLVSNNLGGRWTDSFGGLVFDATGITAPAELK
:
Mb2771|M.bovis_AF2122/97 GKVAFITGAARGLGRAHAVRLA----------------------------
Rv2750|M.tuberculosis_H37Rv GKVAFITGAARGLGRAHAVRLA----------------------------
MUL_3392|M.ulcerans_Agy99 GRVAFITGAARGQGRAHALRLA----------------------------
Mvan_2006|M.vanbaalenii_PYR-1 NKVAVVSGAARGQGRSHAVSLA----------------------------
MAV_2946|M.avium_104 GKVAVISGAARGQGRSHARMLA----------------------------
TH_3929|M.thermoresistible__bu GKVVAITGAARGQGRSHAVHMA----------------------------
MSMEG_4801|M.smegmatis_MC2_155 GKVAFITGAARGQGRSHAVRLA----------------------------
Mflv_4148|M.gilvum_PYR-GCK GKVAFITGAARGQGRAHAIAMA----------------------------
MAB_0072c|M.abscessus_ATCC_199 GKVALVTGAARGIGRAQAVRFA----------------------------
MMAR_4754|M.marinum_M GKVAFITGAARGQGRSHAVRLA----------------------------
MLBr_02565|M.leprae_Br4923 GLHNFFTPLLRNLGRCARVVVIGTTPDATTSTDERIAQRALEGFSRSLGK
. .: *. **. .
Mb2771|M.bovis_AF2122/97 --ADGANIIAVDICEQ-------------------------------IAS
Rv2750|M.tuberculosis_H37Rv --ADGANIIAVDICEQ-------------------------------IAS
MUL_3392|M.ulcerans_Agy99 --RDGADVIAVDLCDQ-------------------------------IAS
Mvan_2006|M.vanbaalenii_PYR-1 --AEGAHIIALDICAD-------------------------------LEG
MAV_2946|M.avium_104 --AEGADIIAVDLCAD-------------------------------IET
TH_3929|M.thermoresistible__bu --DEGADIIALDICAD-------------------------------IAT
MSMEG_4801|M.smegmatis_MC2_155 --EEGADIIAVDLCQD-------------------------------IDS
Mflv_4148|M.gilvum_PYR-GCK --REGADIIAVDICRD-------------------------------IPS
MAB_0072c|M.abscessus_ATCC_199 --QEGADIIALDICAP-------------------------------VHT
MMAR_4754|M.marinum_M --QEGADIIAVDVCKPI------------------------------VEN
MLBr_02565|M.leprae_Br4923 ELRRGATVALVYLSPDAKPAATGLESTMRFILSAKSAYVNGQVFYVGAAD
** : : :.
Mb2771|M.bovis_AF2122/97 VPYPLSTADDLAATVELVEDAGGGIVARQGDVRDRA----------SLSV
Rv2750|M.tuberculosis_H37Rv VPYPLSTADDLAATVELVEDAGGGIVARQGDVRDRA----------SLSV
MUL_3392|M.ulcerans_Agy99 VPYPLGTAEELATTVKLVEDTGARIVASQADVRDRE----------ALAA
Mvan_2006|M.vanbaalenii_PYR-1 NTYPLSRPEDLDETARLAEKEGVRVHTAIVDVRERA----------AVWK
MAV_2946|M.avium_104 NEYPLARPEDLDETARLVEKEGQRAITAVADVRDRV----------VLSA
TH_3929|M.thermoresistible__bu NEYPLATREDLEETARLVEKAGRRAITAEVDVRDRA----------RLKQ
MSMEG_4801|M.smegmatis_MC2_155 IGYSLATPEDLEETARYVEKAGRRIVTAQADVRDAA----------QLKE
Mflv_4148|M.gilvum_PYR-GCK NPYPLATPDDLAETERSVKETGRRVVARVADVRERH----------ELRE
MAB_0072c|M.abscessus_ATCC_199 TITPPATRADLDKTSELVESAGRRVIPGVVDVRNLR----------DVQT
MMAR_4754|M.marinum_M TTIPASTPEDLAETADLVKGHNRRIFTAEADVRDYD----------ALKA
MLBr_02565|M.leprae_Br4923 STPPTNWDRPLENKVAIVTGAARGIGAAIAEVVARDGARVVAIDVESAAE
. * . . . :*
Mb2771|M.bovis_AF2122/97 ALQAGLDEFG---------------------------RLDIVVANAGIAM
Rv2750|M.tuberculosis_H37Rv ALQAGLDEFG---------------------------RLDIVVANAGIAM
MUL_3392|M.ulcerans_Agy99 ALQAGIDELG---------------------------QVDIVVANAGIAP
Mvan_2006|M.vanbaalenii_PYR-1 AVADGVDALG---------------------------RLDIVVANAGICP
MAV_2946|M.avium_104 AIDAGVAEFG---------------------------HLDIVVANAGICP
TH_3929|M.thermoresistible__bu VLDDAVAELG---------------------------GLHVVVANAGICP
MSMEG_4801|M.smegmatis_MC2_155 ALERGVSELG---------------------------KLDIVVAQAGIAA
Mflv_4148|M.gilvum_PYR-GCK AVEAGVVDFG---------------------------KIDIVVANAGILP
MAB_0072c|M.abscessus_ATCC_199 FTQDAVTEFG---------------------------GLDIVCATAGITS
MMAR_4754|M.marinum_M AVDAGVDELG---------------------------RLDIIVANAGIGN
MLBr_02565|M.leprae_Br4923 ALDETANRVGGTTLSLDVTADDAVDKITEHLHDYQGGHADILVNNAGITR
.* .:: ***
Mb2771|M.bovis_AF2122/97 --MQAGDD---GWRDVIDVNLTGVFHTVQVAIPTLIEQGTGGSIVLISSA
Rv2750|M.tuberculosis_H37Rv --MQAGDD---GWRDVIDVNLTGVFHTVQVAIPTLIEQGTGGSIVLISSA
MUL_3392|M.ulcerans_Agy99 --MQSGDD---GWRDVIDVNLSGAYYTVEVAIPTMIEQGRGGSIVLISSA
Mvan_2006|M.vanbaalenii_PYR-1 --MGRGQT-IQSWSDAIDTDLVGVFNVIQASLPHVNDGA---SIIATGSL
MAV_2946|M.avium_104 --LTAGLP-PQAFADAVDVDLGGVLNLVHASLKHLRAGA---SIIVIGSN
TH_3929|M.thermoresistible__bu --QGNHLP-YKAYIDAFDVDFVGVVNTIHVGLDYLGEGG---SVIVTGSI
MSMEG_4801|M.smegmatis_MC2_155 --MKGNPP-LQAWTDGINTNLVGTINAIQVALPHLSEGA---SIVATASA
Mflv_4148|M.gilvum_PYR-GCK --MAMGNPDPMGFVDASDVDLVGVMNTVAVAIPHLPDGA---SVIVTGST
MAB_0072c|M.abscessus_ATCC_199 RRMLLDMP-ESDWQTMLDVNLTGVWHTTRAATPHLIARG-GGAMVLVSSI
MMAR_4754|M.marinum_M GGATLDKTSEHDWQEMIDVNLSGVWKSVKAAVPHLIAGGNGGSIVLTSSV
MLBr_02565|M.leprae_Br4923 DKLLANMD-DCRWDAVLEVNLLAPQRLTEGLVGNGSISK-GGRVIVLSSM
: :.:: . :: .*
Mb2771|M.bovis_AF2122/97 AGLVGIGSS--------DPGSLGYAAAKHGVVGLMRAYANHLAPQNIRVN
Rv2750|M.tuberculosis_H37Rv AGLVGIGSS--------DPGSLGYAAAKHGVVGLMRAYANHLAPQNIRVN
MUL_3392|M.ulcerans_Agy99 AGLVGISSA--------DAGAIGYVASKHALVGLMRVYANLLAPHSIRVN
Mvan_2006|M.vanbaalenii_PYR-1 AAQLGSATNQ-------GPGGSAYSLAKQVVAHYVNDLSIQLAKRMIRVN
MAV_2946|M.avium_104 AAFMSSLNTSGAGSGIGGPGGAGYAFAKLAAAHYVNDFALALAPFSIRMN
TH_3929|M.thermoresistible__bu AGLVDQKDMANGG-GPSGPGGAGYGMAKKMVRDYTKALALTLAPHKIRIN
MSMEG_4801|M.smegmatis_MC2_155 AALMDAHNKPNPG---ADPGGMGYMVSKRMISEYVHYLATELTPLGIRAN
Mflv_4148|M.gilvum_PYR-GCK AGMIRGTTTSPDM----GPGGAGYGWSKRIVIEYIEEMSLQLAPRMIRVN
MAB_0072c|M.abscessus_ATCC_199 AGLRG------------LVGVSHYVAAKHGVVGLMRSLAIELAPHKIRVN
MMAR_4754|M.marinum_M GGMKA------------YPHCGNYVAAKHGVVGLMRSFAVELGQHMIRVN
MLBr_02565|M.leprae_Br4923 AGIAG------------NRGQTNYATAKAGVIGLTQALAPGLAEKGITIN
.. * :* . : * * *
Mb2771|M.bovis_AF2122/97 SVHPCGVDTPMINN-EFFQQWLT----------TADMDAPHNLGNALPVE
Rv2750|M.tuberculosis_H37Rv SVHPCGVDTPMINN-EFFQQWLT----------TADMDAPHNLGNALPVE
MUL_3392|M.ulcerans_Agy99 SLHPSGVDPPMINN-EFIRHWLADL--------VAETGSGPGAGNALPVQ
Mvan_2006|M.vanbaalenii_PYR-1 AIHPTNVNTDMLHNEGLYRVFRPDL----KEPTREEAEEAFPAMQAMPIP
MAV_2946|M.avium_104 AVHPTNVNTDMLHSPPMYRAFRPDL----PAPTREDAEPVFPLVQAMPVP
TH_3929|M.thermoresistible__bu AVHPTNVNTDMLHNMSMYRVFRPDL----TEPTREDAEVVFPLLQAMPIP
MSMEG_4801|M.smegmatis_MC2_155 VIHPTNCNTDMLQSEPMYRSFRPDL----EHPTRADAEPVFYVQQAMKVP
Mflv_4148|M.gilvum_PYR-GCK GIHPTNCNTHLLQNEGMYGVFRPDLKMEGKTPTREDAEPLFSLFQAMPIP
MAB_0072c|M.abscessus_ATCC_199 TVHPTNVDTPMIQNEFVRTLFRPDL----EQPTREDFAAAAKTMNLLDLP
MMAR_4754|M.marinum_M SVHPTHVRTPMLHNEGTFKMFRPDL----ENPGPDDMAPICQLFHTLPIP
MLBr_02565|M.leprae_Br4923 AVAPGFIETQMTAAIPLAT--------------REVGRRLNSLLQGG---
: * . : :
Mb2771|M.bovis_AF2122/97 LVQPTDIANAVAWLASEEARYVTGVTLPVDAGFVNKR------
Rv2750|M.tuberculosis_H37Rv LVQPTDIANAVAWLASEEARYVTGVTLPVDAGFVNKR------
MUL_3392|M.ulcerans_Agy99 ILQADDIAGALAWLVSDEARYITGVALPVDAGCVNKR------
Mvan_2006|M.vanbaalenii_PYR-1 YIEPRDVSNAVVFLAGDDSRYITGTQLRIDAGGYVKAVPWKG-
MAV_2946|M.avium_104 YVEPEDISEAVLFLASDAARYITGQQLRVDAGGFLKVKPWSGG
TH_3929|M.thermoresistible__bu YVEPDDVSHAVVYLASDESRYVTGQQLFVDAGAGLKMGV----
MSMEG_4801|M.smegmatis_MC2_155 WVEPSDISNAVLWLASDESRFVTGMQLRVDAGGYLKWYDYHV-
Mflv_4148|M.gilvum_PYR-GCK YIEPEDMANLGVFLASDDSRYITGQQIRVDAGSLLKWPNGLGG
MAB_0072c|M.abscessus_ATCC_199 WVEPLDIANASLFLASDEARYITSVSLPVDAGSTQR-------
MMAR_4754|M.marinum_M WVEAEDISNAVLFLASDESRYITGVTLPVDAGGCLK-------
MLBr_02565|M.leprae_Br4923 --LPVDVAETIAYFAIPASNAVTGNVIRVCGQAMLGA------
. *:: ::. :. :*. : : .