For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTAPQRRGLNDVVTRMWSFLAPAYDLPFLQQWVYRPPHNEVIAQLRNHGSRRIADIACGTGILSERIQRE LNPDEIYGVDMSDGMLNQARAKSDRVQWLRAPAEQLPFDDGALDAVVTTSAFHFFDQPAALREFHRVLAP GGLVAVSALSTRRPLLQASADSRWKPQHNASAPEMRKLFEDAGFVIGDQYRIPRPAWTRLVSDLLTVGTK RD
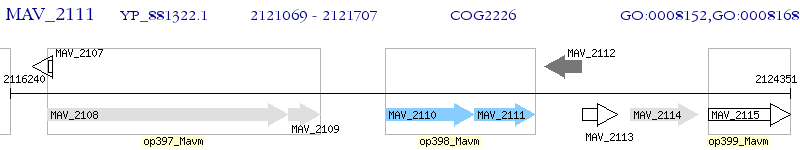
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2111 | - | - | 100% (212) | methyltransferase, UbiE/COQ5 family protein |
| M. avium 104 | MAV_4317 | - | 1e-10 | 35.19% (108) | putative methyltransferase |
| M. avium 104 | MAV_5229 | - | 2e-09 | 36.46% (96) | methyltransferase type 11 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3374 | - | 1e-11 | 37.96% (108) | methyltransferase (methylase) |
| M. gilvum PYR-GCK | Mflv_4877 | - | 5e-11 | 37.62% (101) | methyltransferase type 11 |
| M. tuberculosis H37Rv | Rv3342 | - | 1e-11 | 37.96% (108) | methyltransferase (methylase) |
| M. leprae Br4923 | MLBr_00130 | - | 1e-10 | 33.96% (106) | hypothetical protein MLBr_00130 |
| M. abscessus ATCC 19977 | MAB_4613 | - | 2e-59 | 57.14% (210) | similarity with UbiE/COQ5 methyltransferase |
| M. marinum M | MMAR_3533 | - | 2e-95 | 79.05% (210) | methyltransferase (methylase) |
| M. smegmatis MC2 155 | MSMEG_0098 | - | 3e-11 | 37.93% (116) | methyltransferase |
| M. thermoresistible (build 8) | TH_1968 | - | 3e-10 | 37.04% (108) | POSSIBLE METHYLTRANSFERASE (METHYLASE) |
| M. ulcerans Agy99 | MUL_2765 | - | 6e-95 | 79.05% (210) | methyltransferase (methylase) |
| M. vanbaalenii PYR-1 | Mvan_5517 | - | 2e-12 | 31.43% (175) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3533|M.marinum_M -----MTTVP-----------RRGFNDAVTGIWS----------------
MUL_2765|M.ulcerans_Agy99 -----MTTVP-----------RRGFNDAVTGIWS----------------
MAV_2111|M.avium_104 -----MTAPQ-----------RRGLNDVVTRMWS----------------
MAB_4613|M.abscessus_ATCC_1997 -----MTTADGLHSGDPSPVHRRAFNDQVTRFWG----------------
Mb3374|M.bovis_AF2122/97 -----MTCSR----------------RDMSLSFG----------------
Rv3342|M.tuberculosis_H37Rv -----MTCSR----------------RDMSLSFG----------------
Mflv_4877|M.gilvum_PYR-GCK -----MTAES----------------DRRSLSFG----------------
TH_1968|M.thermoresistible__bu ------------------------------LSFG----------------
Mvan_5517|M.vanbaalenii_PYR-1 -----MVSMD---------HMTENPPHPVARTWGNRHDYL----PAAGRD
MSMEG_0098|M.smegmatis_MC2_155 MDTALRRAMDLLIDPPAEPDFSKGYLDLLGRGDSTAPKNTGLIQKAWASP
MLBr_00130|M.leprae_Br4923 ---MAFTRIHS-------FLASAGNTSMYKRVWRFWYPLMTHKLGTDEIM
MMAR_3533|M.marinum_M LLAPAYDLAILQQWVYRPPHDEVLAQLRAHNS---------RRIADIACG
MUL_2765|M.ulcerans_Agy99 LLAPAYDLAILQQWVYRPPHDEVLAQLRAHKS---------RRIADIACG
MAV_2111|M.avium_104 FLAPAYDLPFLQQWVYRPPHNEVIAQLRNHGS---------RRIADIACG
MAB_4613|M.abscessus_ATCC_1997 RAARAYDWAPLQQWVYRPAQDEMLAQLRQHKS---------QRIVDIACG
Mb3374|M.bovis_AF2122/97 SAVGAYERGRPS---YPPEAIDWLLPAAARR------------VLDLGAG
Rv3342|M.tuberculosis_H37Rv SAVGAYERGRPS---YPPEAIDWLLPAAARR------------VLDLGAG
Mflv_4877|M.gilvum_PYR-GCK AEAAAYERGRPS---YPPEAIDWLLADSGSRR---------LEVLDLGAG
TH_1968|M.thermoresistible__bu AEAAAYERGRPS---YPPEAIDWLLP-SDAR-----------DVLDLGAG
Mvan_5517|M.vanbaalenii_PYR-1 FFLPAYDLLVRLLG-TYPLYDELVTQAELAGE---------LDVLEIGCG
MSMEG_0098|M.smegmatis_MC2_155 LGSKLYDRAQLLNRRLVASTRPPVEWLRIPPG---------GTVLDIGCG
MLBr_00130|M.leprae_Br4923 FINWAYEEDPPMALPLEASDEPNRAHINLYHRTATQVNLSGKRILEVSCG
*: . : ::..*
MMAR_3533|M.marinum_M TGILSDRIERELHPDEIYG-VDMSHGMLDQARAKSD---RVRWLRGPAEQ
MUL_2765|M.ulcerans_Agy99 TGILSARIERELHPDEIYG-VDMSHGMLDQARAKSD---RVRWLRGPAEQ
MAV_2111|M.avium_104 TGILSERIQRELNPDEIYG-VDMSDGMLNQARAKSD---RVQWLRAPAEQ
MAB_4613|M.abscessus_ATCC_1997 TGILATRIQDELEPEQVHG-VDMSEGMLAQAKARSS---LVNWQFAPAEK
Mb3374|M.bovis_AF2122/97 TGKLTTRLVERG--LDVVA-VDPIPEMLDVLRAALP---QTVALLGTAEE
Rv3342|M.tuberculosis_H37Rv TGKLTTRLVERG--LDVVA-VDPIPEMLDVLRAALP---QTVALLGTAEE
Mflv_4877|M.gilvum_PYR-GCK TGKLTTRLVERG--LDVVA-VDPIPEMLDLLRTSLP---DTPALLGTAEE
TH_1968|M.thermoresistible__bu TGKLTTRLVERG--LNVTA-VDPIPEMLELLSKSLP---DTPALLGTAEE
Mvan_5517|M.vanbaalenii_PYR-1 TGNLTARALRAAPSARITA-TDPDPRAVTRARRKAAGEGPVRFETAYAQE
MSMEG_0098|M.smegmatis_MC2_155 PGNITAQLARAAGLDGLALGVDISEPMLARAVAAEAG-RQVGFVRADAQQ
MLBr_00130|M.leprae_Br4923 HGGGASYLTRALHPASYTG-LDLNPAGIKLCQKRHQLP-GLEFVRGDAEN
* : * : . *::
MMAR_3533|M.marinum_M LPFDDGALDAVVTTTAFHFFDQ---PAALREFHRVLAPGGLVAVSSLS--
MUL_2765|M.ulcerans_Agy99 LPFDDGALDAVVTITAFHFFDQ---PAALREFHRVLAPGGLVAVSSLS--
MAV_2111|M.avium_104 LPFDDGALDAVVTTSAFHFFDQ---PAALREFHRVLAPGGLVAVSALS--
MAB_4613|M.abscessus_ATCC_1997 LPFDDGALDAVVSTSAFHFFDQ---PAALTEFHRVLTPGGFAAITTFTPD
Mb3374|M.bovis_AF2122/97 IPLDDNSVDAVLVAQAWHWVDP---ARAIPEVARVLRPGGRLGLVWNTRD
Rv3342|M.tuberculosis_H37Rv IPLDDNSVDAVLVAQAWHWVDP---ARAIPEVARVLRPGGRLGLVWNTRD
Mflv_4877|M.gilvum_PYR-GCK IPLPDDSVDAVLVAQAWHWFDE---TRAVKEIARVLRPGGSLGLVWNNRD
TH_1968|M.thermoresistible__bu IPLPDNSVDAVLVAQAWHWFDV---DRAVREVARVLRPGGRLGLVWNTRD
Mvan_5517|M.vanbaalenii_PYR-1 LPFADASFDRVLSSLMLHHLDDGTKVSALAEAWRVLRPGGRLHIVDVG--
MSMEG_0098|M.smegmatis_MC2_155 LPFRDEVFDAATSLAVFQLIPDP--VAAVSEIVRVLKPGGRVAIMVPT--
MLBr_00130|M.leprae_Br4923 LPFDNESFDVVINIEASHCYPH--FPRFLAEVVRVLRPGGHLAYADLRPS
:*: : .* . : : * *** ***
MMAR_3533|M.marinum_M ------------------------ARQPLLQLPAASKWKPQHSPSPAEMR
MUL_2765|M.ulcerans_Agy99 ------------------------ARQPLLQLPAASKWKPQHSPSPAEMR
MAV_2111|M.avium_104 ------------------------TRRPLLQASADSRWKPQHNASAPEMR
MAB_4613|M.abscessus_ATCC_1997 R-----------------------TSAPILRRVFGDR-IPASVPSKAEMR
Mb3374|M.bovis_AF2122/97 ERLGWVRELGEIIGRDGDPVRDRVTLPEPFTTVQRHQVEWTNYLTPQALI
Rv3342|M.tuberculosis_H37Rv ERLGWVRELGEIIGRDGDPVRDRVTLPEPFTTVQRHQVEWTNYLTPQALI
Mflv_4877|M.gilvum_PYR-GCK ERLGWVKDLGDIIGHEGDPWSQSVELPAPFTGVERHHVEWTSYLTPQALI
TH_1968|M.thermoresistible__bu ERMGWVKELGRIIGDEHDPLSKTVTLPEPFTDVQRHQVEWTNYLTPQALI
Mvan_5517|M.vanbaalenii_PYR-1 -------------------------GADQPEGLMSRLTGHRHAAAGARLP
MSMEG_0098|M.smegmatis_MC2_155 --------------------------AGAVKPVTFLARGGARIFGDDELG
MLBr_00130|M.leprae_Br4923 NKVG--EWEVDFANSRLQQLSQREINAEVLRGIASNSQKSRD-LVDRHLP
:
MMAR_3533|M.marinum_M ALFEGAGFTISDQHRVRRPVWTRLVP----------------DLITVGAK
MUL_2765|M.ulcerans_Agy99 ALFEGAGFTISDQHRVRRPVWTRLVP----------------DLITVGTK
MAV_2111|M.avium_104 KLFEDAGFVIGDQYRIPRPAWTRLVS----------------DLLTVGTK
MAB_4613|M.abscessus_ATCC_1997 LMFETAGFEVTVQRPVHHPFTLPFIEF---------------DRLTVGIK
Mb3374|M.bovis_AF2122/97 DLVASRSYCITSPAQVRTKTLDRVRQLLATHPALANSNGLALPYVTVCVR
Rv3342|M.tuberculosis_H37Rv DLVASRSYCITSPAQVRTKTLDRVRQLLATHPALANSNGLALPYVTVCVR
Mflv_4877|M.gilvum_PYR-GCK DLVASRSYCITSPERVRARTLDRVRELLTTHPALANSSGLALPYVTVCVR
TH_1968|M.thermoresistible__bu DLVASRSYCITSPAKVRARTLERVRELLATHPALANNSGLALPYITVCIR
Mvan_5517|M.vanbaalenii_PYR-1 QLVEAAGFDTTVVATRRLRVFG------------------PVSFIRATRP
MSMEG_0098|M.smegmatis_MC2_155 DIFENVGLVGVRVKTHGFIQWVRGTKL-----------------------
MLBr_00130|M.leprae_Br4923 AFLRFAGREFIGVQGTQLSRYLEGGE---------------LSYRMYSFA
:. .
MMAR_3533|M.marinum_M S---
MUL_2765|M.ulcerans_Agy99 S---
MAV_2111|M.avium_104 RD--
MAB_4613|M.abscessus_ATCC_1997 R---
Mb3374|M.bovis_AF2122/97 ATLA
Rv3342|M.tuberculosis_H37Rv ATLA
Mflv_4877|M.gilvum_PYR-GCK ATLA
TH_1968|M.thermoresistible__bu ATLA
Mvan_5517|M.vanbaalenii_PYR-1 GE--
MSMEG_0098|M.smegmatis_MC2_155 ----
MLBr_00130|M.leprae_Br4923 KD--