For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RIGVVFPQTEIGSDPAVLRSYAQRVEELGFTHILAYDHVAGADPRVHRGWRGPYDIDSTFHEPFVLFGYL AAITSLELVTGVIILPQRQSVLVAKQAAEVDLLTGGRFRLGIGLGWNAVEYEALGETFTNRGKRSEEQVE LMRRLWTERSVTFDGKYHTVTAAGLAPLPVQRPIPVWFGAASDRAFERAGRLGDGWFPMTEPGPGLDHAL EQVTRAAEAAGRDPTSLGMEGRVSWTDDRDKLAGDIAAWRAAGATHLSVNTMKAGFAGVDEHLAALERVA ADLR*
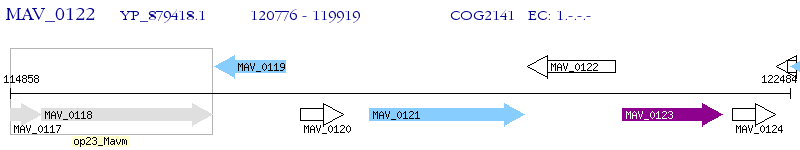
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0122 | - | - | 100% (285) | hypothetical protein MAV_0122 |
| M. avium 104 | MAV_2935 | - | 1e-29 | 35.81% (229) | hypothetical protein MAV_2935 |
| M. avium 104 | MAV_2659 | - | 1e-27 | 34.15% (205) | hypothetical protein MAV_2659 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2185c | - | 3e-22 | 35.11% (225) | hypothetical protein Mb2185c |
| M. gilvum PYR-GCK | Mflv_4839 | - | 1e-30 | 37.22% (223) | luciferase family protein |
| M. tuberculosis H37Rv | Rv2161c | - | 3e-22 | 35.11% (225) | hypothetical protein Rv2161c |
| M. leprae Br4923 | MLBr_00131 | - | 1e-13 | 29.49% (234) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_0366 | - | 7e-31 | 34.59% (292) | hypothetical protein MAB_0366 |
| M. marinum M | MMAR_1586 | - | 2e-28 | 32.65% (245) | oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_2811 | - | 4e-33 | 36.12% (227) | hydride transferase 1 |
| M. thermoresistible (build 8) | TH_2146 | - | 1e-28 | 30.69% (290) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_3448 | - | 3e-28 | 31.52% (257) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_5752 | - | 1e-119 | 70.21% (282) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_0122|M.avium_104 ---MRIGVVFP-QTEIGSDPAVLRSYAQRVEELGFTHILAYDHVAGADPR
Mvan_5752|M.vanbaalenii_PYR-1 ---MRIGVVFP-QTELGGDPGAVRAYGRRVEELGFTHLLAYDHVVGADPA
MAB_0366|M.abscessus_ATCC_1997 ---MKIGVVAPVELGVTAEPDKILEFARAAERLGFSEISVVEHAVVIGDT
MMAR_1586|M.marinum_M ---MQLGLHAL-GIGAGADRSMIDAVACAADDAGFATLWAGEHVVMVDRH
MUL_3448|M.ulcerans_Agy99 ---MQLGLHAL-EIGAGADRSMIDAVACAADDAGFATLWAGEHVVMVDRH
MSMEG_2811|M.smegmatis_MC2_155 ---MQISLHAL-GIGAGARREIIDAVAVAAERAGFTTLWCGEHVVMVDRP
Mflv_4839|M.gilvum_PYR-GCK ---------------------MIDAVAASAERSGFETLWFGDQVVMADED
TH_2146|M.thermoresistible__bu ---MRFAFTYP-MDSHPYHPDLATGAAVARVAATAETAGFSG-FGFTDHP
Mb2185c|M.bovis_AF2122/97 ------MLVSL-MQFVTDLTPPPQLVAVWAEERGFAGLYVPEKTHVPISR
Rv2161c|M.tuberculosis_H37Rv ------MLVSL-MQFVTDLTPPPQLVAVWAEERGFAGLYVPEKTHVPISR
MLBr_00131|M.leprae_Br4923 MAGFRFGFVDALVHTLFPPSLPARASIVSGAVLGADSYWVGDHLNALVPR
MAV_0122|M.avium_104 -VHRGWRGP-----YDIDSTFHEPFVLFGYLAAITS-LELVTGVIILPQ-
Mvan_5752|M.vanbaalenii_PYR-1 -VHTGWSGP-----YDVDTTFHEPLVMFGYLAAVTTTLELVTGVIILPQ-
MAB_0366|M.abscessus_ATCC_1997 -QSTYPYSPTGQSHLPDDIDIPDPLELLSFVAAATTTLGLSTGVLVLPD-
MMAR_1586|M.marinum_M -TSRYPYSEDGVIAIPPQADWLDPMIALSFAAAASSRITVATGVLLLPE-
MUL_3448|M.ulcerans_Agy99 -TSRYPYSEDGVIAIPPQADWLDPMIALSFAAASSSRITVATGVLLLPE-
MSMEG_2811|M.smegmatis_MC2_155 -RSRYPYSADGRIAVAADADWLDPMIALSFAAAATSTIRIATGVLLLPE-
Mflv_4839|M.gilvum_PYR-GCK -GPHHPYRDDGEIPASAQTDWLDPLIALSFAAAATNRIELATGVLVLPE-
TH_2146|M.thermoresistible__bu -APTQKWLDHG------GHDTLDPFVAMGFAAAHTTTLRLIPHIVVLPY-
Mb2185c|M.bovis_AF2122/97 -STPWPGGELP----DWYRRCYDPVVALAAAAAVTTRLRVGTGACLVAV-
Rv2161c|M.tuberculosis_H37Rv -STPWPGGELP----DWYRRCYDPVVALAAAAAVTTRLRVGTGACLVAV-
MLBr_00131|M.leprae_Br4923 SVATPKYLGVAAKVVPKIDANYEPWTMLGNLAAGNRLNGLRLGVCVTDAG
:* :. ** . : :
MAV_0122|M.avium_104 -RQSVLVAKQAAEVDLLTGGRFRLGIGLGWNAVEYEALGETFTNRGKRSE
Mvan_5752|M.vanbaalenii_PYR-1 -RQTALVAKQAAEVDLLSGGRLRFGVGVGWNAVEYEALGEQFTNRGTRSE
MAB_0366|M.abscessus_ATCC_1997 -HHPVVLAKRLATLDRLSRGRLRICAGVGWMREEIEACGGDFDRRGRAAD
MMAR_1586|M.marinum_M -HNPVVVAKQAASLDRLSGGRLTLGVGVGWSREEFAALGVPFQDRAGRTA
MUL_3448|M.ulcerans_Agy99 -HNPVVVAKQAASLDRLSGGRLTLGVGVGWSREEFAALGVPFQDRVGRTA
MSMEG_2811|M.smegmatis_MC2_155 -HNPVIMAKQAASVDQLSGGRLLLGVGIGWSREEFEALGVPFARRGARTA
Mflv_4839|M.gilvum_PYR-GCK -RNPVVTAKQAASLDRMTGGRLTLGVGIGISPQEYDALGVPFASRGARLA
TH_2146|M.thermoresistible__bu -RNPFVVAKAGASVDLLSGGRFTLGVGVGYLKREFAALGVDFEERAALFE
Mb2185c|M.bovis_AF2122/97 -HDPILLAKQIASLCAMSGERFVLGVGFGWNVEELADHGVPFADRIAVTV
Rv2161c|M.tuberculosis_H37Rv -HDPILLAKQIASLCAMSGERFVLGVGFGWNVEELADHGVPFADRIAVTV
MLBr_00131|M.leprae_Br4923 RRNPAVTAQAAATLHLLTRGKAMLGIGVGE-REGNEPYGVEWTKPVARFQ
:.. : *: * : :: : : *.* * :
MAV_0122|M.avium_104 EQVELMRRLWTER--SVTFDGKYHTVTAAGLAPLPVQRP-IPVWFGAASD
Mvan_5752|M.vanbaalenii_PYR-1 EQVELLRQLWTQR--TVTFDGRHHRVTGAGIAPLPVQRP-IPLWIGAAAP
MAB_0366|M.abscessus_ATCC_1997 EAIAVMRALWSTDG-PTSHDGEFYSFRNAYSYPKPVRPQGVPLYIGGHSK
MMAR_1586|M.marinum_M EYVAAMRTLWRDD--LASFTGEFVRFDSVRVNPKPVRDRRVPVMVGGNSD
MUL_3448|M.ulcerans_Agy99 EYVAAMRTLWRDD--LASFTGEFVRFDSVRVNPKPVRDRRVPVMVGGNSD
MSMEG_2811|M.smegmatis_MC2_155 EYVEAMRTLWRQD--VADFTGEFAAFDAVRLHPKPSAG-SIPIIVGGNGD
Mflv_4839|M.gilvum_PYR-GCK EYVGAMRTLWRDD--DASFAGRYVAFDGVRANPKPQGR-SIPVVLGGNSD
TH_2146|M.thermoresistible__bu EALEVIRAIWTTD--NVSYTGRHFTADGITAHPHPVTKPHPPIWIGGNTG
Mb2185c|M.bovis_AF2122/97 DKLAAMRALWAAE--PVHYEGTHASVPPSWAWPKPAVA--PPVLFGCRPS
Rv2161c|M.tuberculosis_H37Rv DKLAAMRALWAAE--PVHYEGTHASVPPSWAWPKPAVA--PPVLFGCRPS
MLBr_00131|M.leprae_Br4923 EALATIRALWDSNGELVSRESQFFPLHNALFDLPPYRGKWPEIWVAAHGP
: : :* :* . . . * : ..
MAV_0122|M.avium_104 R-AFERAGRLGDGWFPMTE--------------PGPGLDHALEQVTRAAE
Mvan_5752|M.vanbaalenii_PYR-1 R-GYARAGRIADGWFPLVG--------------PGPALDEAKGAVDAAAR
MAB_0366|M.abscessus_ATCC_1997 A-AARRAGRLGSGFQPLGL--------------VGDDLTAAVGVMRAAAV
MMAR_1586|M.marinum_M A-ALRRVAAWADGWYGFN-------------LDGVGAVRERVGTLDRLCA
MUL_3448|M.ulcerans_Agy99 A-ALRRVAAWADGWYGFN-------------LDGVGAVRERVGTLDRLCA
MSMEG_2811|M.smegmatis_MC2_155 R-ALARVASWGDGWYGFN-------------LDGVAAVGERMDMLRRLCA
Mflv_4839|M.gilvum_PYR-GCK A-ALRRVAEWGDGWYGFD-------------LADVEEAAFRASKLRSMCR
TH_2146|M.thermoresistible__bu A-ARQRVADHGAGWCPFPAPAVLAQTARTAAMDSAEALAAGITDLHRRLE
Mb2185c|M.bovis_AF2122/97 ARAFEVIARHGDGWQPIEG---------------YGELLGALPMLHAAFE
Rv2161c|M.tuberculosis_H37Rv ARAFEVIARHGDGWQPIEG---------------YGELLGALPMLHAAFE
MLBr_00131|M.leprae_Br4923 R-MLQATGRYADAWIPIVLVR-------------PTDYSCALEVVRTAAS
. . .: : :
MAV_0122|M.avium_104 AAGRDPTSLGMEGRVSWTDDRDKLAGDIAAWRAAG------------ATH
Mvan_5752|M.vanbaalenii_PYR-1 EAGRDPSEIGMEGRVRWRHDLDQTAADIGEWAEHG------------ASH
MAB_0366|M.abscessus_ATCC_1997 DAGRDPRGLDLVLGHGLHR-VDQAALDQAAHAGAG------------RML
MMAR_1586|M.marinum_M ESGRNRDELRLSVA---LRCPQPCDVDALAELGID-------------EL
MUL_3448|M.ulcerans_Agy99 ESGRNRDELRLSVA---LRCPQPCDVDALAELGID-------------EL
MSMEG_2811|M.smegmatis_MC2_155 RAGRDVDDLHCAVA---LQAPAVGDIPALVDLGVD-------------EL
Mflv_4839|M.gilvum_PYR-GCK EFGRDPTELKLSVA---LHDPVPADAVRLADAGID-------------EL
TH_2146|M.thermoresistible__bu ATGRDPADVDVVFSN--LTGGTPGRADFNADAYLDGVAKL---GALGVTW
Mb2185c|M.bovis_AF2122/97 RAGRDPATAQVCVY---SSAGDPATLHEYRRAGVA-----------EVAL
Rv2161c|M.tuberculosis_H37Rv RAGRDPATAQVCVY---SSAGDPATLHEYRRAGVA-----------EVAL
MLBr_00131|M.leprae_Br4923 DAGRDPMSITPAAVRGIITGRTRDDVDEALDSVLVRMIALGVPGEAWARH
**:
MAV_0122|M.avium_104 LSVNTMKAGFAGVDEHLAALERVAADLR----------------------
Mvan_5752|M.vanbaalenii_PYR-1 LSVDTMRAGLRTVNEHLAVLEAIADRVGLR--------------------
MAB_0366|M.abscessus_ATCC_1997 LSASRRATTLEAVLEEMAGCATRLELGGPR--------------------
MMAR_1586|M.marinum_M VLVETPPEAADAAQNWVSALAERWLSTVWVR-------------------
MUL_3448|M.ulcerans_Agy99 VLVETPPEAADAAQNWVSALAERWLSTVWVR-------------------
MSMEG_2811|M.smegmatis_MC2_155 VIVESPPQDPDVAADWVDALADRWITCI----------------------
Mflv_4839|M.gilvum_PYR-GCK VLVAGPPADPIAAEAWIADLAGQWMRVRT---------------------
TH_2146|M.thermoresistible__bu LQVPIPGDSLNHALEAIEAFGSEVIGKL----------------------
Mb2185c|M.bovis_AF2122/97 ALPSAGRDQVLAALDRSAPLVDAFAGDDREVKSHA---------------
Rv2161c|M.tuberculosis_H37Rv ALPSAGRDQVLAALDRSAPLVDAFAGDDREVKSHA---------------
MLBr_00131|M.leprae_Br4923 GVEHPMGADFAGVQDIIPQTIDEETVVSYAAKVPAALMKEVLFSGTPEEV
.
MAV_0122|M.avium_104 -----------------------------------------------
Mvan_5752|M.vanbaalenii_PYR-1 -----------------------------------------------
MAB_0366|M.abscessus_ATCC_1997 -----------------------------------------------
MMAR_1586|M.marinum_M -----------------------------------------------
MUL_3448|M.ulcerans_Agy99 -----------------------------------------------
MSMEG_2811|M.smegmatis_MC2_155 -----------------------------------------------
Mflv_4839|M.gilvum_PYR-GCK -----------------------------------------------
TH_2146|M.thermoresistible__bu -----------------------------------------------
Mb2185c|M.bovis_AF2122/97 -----------------------------------------------
Rv2161c|M.tuberculosis_H37Rv -----------------------------------------------
MLBr_00131|M.leprae_Br4923 IDQVAEWRDHGLKYLVVINGSLVNSSLRKTVSALLPHARVLRGLKKL