For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VKIGVVAPVELGVTAEPDKILEFARAAERLGFSEISVVEHAVVIGDTQSTYPYSPTGQSHLPDDIDIPDP LELLSFVAAATTTLGLSTGVLVLPDHHPVVLAKRLATLDRLSRGRLRICAGVGWMREEIEACGGDFDRRG RAADEAIAVMRALWSTDGPTSHDGEFYSFRNAYSYPKPVRPQGVPLYIGGHSKAAARRAGRLGSGFQPLG LVGDDLTAAVGVMRAAAVDAGRDPRGLDLVLGHGLHRVDQAALDQAAHAGAGRMLLSASRRATTLEAVLE EMAGCATRLELGGPR
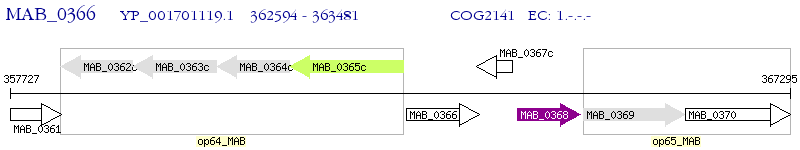
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0366 | - | - | 100% (295) | hypothetical protein MAB_0366 |
| M. abscessus ATCC 19977 | MAB_4908c | - | 3e-21 | 34.50% (200) | putative luciferase-like oxidoreductase |
| M. abscessus ATCC 19977 | MAB_2449 | - | 7e-21 | 29.44% (214) | hypothetical protein MAB_2449 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3106c | - | 3e-27 | 35.19% (216) | hypothetical protein Mb3106c |
| M. gilvum PYR-GCK | Mflv_4839 | - | 7e-33 | 34.85% (264) | luciferase family protein |
| M. tuberculosis H37Rv | Rv3079c | - | 3e-27 | 35.19% (216) | hypothetical protein Rv3079c |
| M. leprae Br4923 | MLBr_00131 | - | 5e-17 | 31.61% (193) | putative oxidoreductase |
| M. marinum M | MMAR_1586 | - | 5e-40 | 39.55% (268) | oxidoreductase |
| M. avium 104 | MAV_2935 | - | 3e-83 | 53.82% (288) | hypothetical protein MAV_2935 |
| M. smegmatis MC2 155 | MSMEG_2811 | - | 5e-39 | 40.97% (227) | hydride transferase 1 |
| M. thermoresistible (build 8) | TH_2945 | - | 2e-31 | 36.18% (246) | PUTATIVE luciferase family protein |
| M. ulcerans Agy99 | MUL_3448 | - | 2e-40 | 39.55% (268) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_5752 | - | 5e-36 | 36.59% (287) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3106c|M.bovis_AF2122/97 -------MQFGVLTFVTDEGIGPAELGAALEHRGFESLFLAEHTHIPVNT
Rv3079c|M.tuberculosis_H37Rv -------MQFGVLTFVTDEGIGPAELGAALEHRGFESLFLAEHTHIPVNT
MAB_0366|M.abscessus_ATCC_1997 ---MKIGVVAPVELGVTAEPDKILEFARAAERLGFSEISVVEHAVVIGDT
MAV_2935|M.avium_104 --------MAPVADGVTADPDWMVAFARHLEACGFESIVAVEHTVLLTRY
MMAR_1586|M.marinum_M ----MQLGLHALGIGAGADRSMIDAVACAADDAGFATLWAGEHVVMVDRH
MUL_3448|M.ulcerans_Agy99 ----MQLGLHALEIGAGADRSMIDAVACAADDAGFATLWAGEHVVMVDRH
MSMEG_2811|M.smegmatis_MC2_155 ----MQISLHALGIGAGARREIIDAVAVAAERAGFTTLWCGEHVVMVDRP
Mflv_4839|M.gilvum_PYR-GCK ---------------------MIDAVAASAERSGFETLWFGDQVVMADED
Mvan_5752|M.vanbaalenii_PYR-1 ----MRIGVVFPQTELGGDPGAVRAYGRRVEELGFTHLLAYDHVVGADP-
TH_2945|M.thermoresistible__bu ------MKFTFALPHMMELDAMMQPWEMAVTGADQTRMARRAEELGFDMI
MLBr_00131|M.leprae_Br4923 MAGFRFGFVDALVHTLFPPSLPARASIVSGAVLGADSYWVGDHLNALVPR
. .
Mb3106c|M.bovis_AF2122/97 QSPYPG---GGPIPEKYYRT-LDPFVALAAAAATTQSLVLGTGIALIPE-
Rv3079c|M.tuberculosis_H37Rv QSPYPG---GGPIPEKYYRT-LDPFVALAAAAATTQSLVLGTGIALIPE-
MAB_0366|M.abscessus_ATCC_1997 QSTYPYSPTGQSHLPDDIDI-PDPLELLSFVAAATTTLGLSTGVLVLPD-
MAV_2935|M.avium_104 DSVYPYDSSGRVGLAPDCPI-PDPLDLLAFLAGHTTRLGLATGVLVLPN-
MMAR_1586|M.marinum_M TSRYPYSEDGVIAIPPQADW-LDPMIALSFAAAASSRITVATGVLLLPE-
MUL_3448|M.ulcerans_Agy99 TSRYPYSEDGVIAIPPQADW-LDPMIALSFAAASSSRITVATGVLLLPE-
MSMEG_2811|M.smegmatis_MC2_155 RSRYPYSADGRIAVAADADW-LDPMIALSFAAAATSTIRIATGVLLLPE-
Mflv_4839|M.gilvum_PYR-GCK GPHHPYRDDGEIPASAQTDW-LDPLIALSFAAAATNRIELATGVLVLPE-
Mvan_5752|M.vanbaalenii_PYR-1 ----AVHTGWSGPYDVDTTF-HEPLVMFGYLAAVTTTLELVTGVIILPQ-
TH_2945|M.thermoresistible__bu AVPEHIVMPREHVDLSGGHW-LHSTIAQAYFAGATERITLNSCVTILPL-
MLBr_00131|M.leprae_Br4923 SVATPKYLGVAAKVVPKIDANYEPWTMLGNLAAGNRLNGLRLGVCVTDAG
.. . *. . : : :
Mb3106c|M.bovis_AF2122/97 -RDPIVTAKEVASLDLVSQGRFRFGVGVGWLREEVANHGVDPAVRGRVID
Rv3079c|M.tuberculosis_H37Rv -RDPIVTAKEVASLDLVSQGRFRFGVGVGWLREEVANHGVDPAVRGRVID
MAB_0366|M.abscessus_ATCC_1997 -HHPVVLAKRLATLDRLSRGRLRICAGVGWMREEIEACGGDFDRRGRAAD
MAV_2935|M.avium_104 -HHPVVLAKRVGTLDALSGGRLRLCVGVGWLQEELRACGADFDNRGRRAD
MMAR_1586|M.marinum_M -HNPVVVAKQAASLDRLSGGRLTLGVGVGWSREEFAALGVPFQDRAGRTA
MUL_3448|M.ulcerans_Agy99 -HNPVVVAKQAASLDRLSGGRLTLGVGVGWSREEFAALGVPFQDRVGRTA
MSMEG_2811|M.smegmatis_MC2_155 -HNPVIMAKQAASVDQLSGGRLLLGVGIGWSREEFEALGVPFARRGARTA
Mflv_4839|M.gilvum_PYR-GCK -RNPVVTAKQAASLDRMTGGRLTLGVGIGISPQEYDALGVPFASRGARLA
Mvan_5752|M.vanbaalenii_PYR-1 -RQTALVAKQAAEVDLLSGGRLRFGVGVGWNAVEYEALGEQFTNRGTRSE
TH_2945|M.thermoresistible__bu -QHPLVTAKALATADWLSGGRMMVTFGVGWLQREFELLGVPFRKRGRIAD
MLBr_00131|M.leprae_Br4923 RRNPAVTAQAAATLHLLTRGKAMLGIGVGE-REGNEPYGVEWTKPVARFQ
:.. : *: . . :: *: . *:* *
Mb3106c|M.bovis_AF2122/97 ERLRAIIEIWTQ---EQAEFHGTYVDFDPIYCWPKPVTKPYPPLYVGG-G
Rv3079c|M.tuberculosis_H37Rv ERLRAIIEIWTQ---EQAEFHGTYVDFDPIYCWPKPVTKPYPPLYVGG-G
MAB_0366|M.abscessus_ATCC_1997 EAIAVMRALWST--DGPTSHDGEFYSFRNAYSYPKPVRPQGVPLYIGGHS
MAV_2935|M.avium_104 EQLAVLRALWAHRPDG-ASFHGEFFTFDNVMCYPKPVAAERFPVHIGGHS
MMAR_1586|M.marinum_M EYVAAMRTLWRD---DLASFTGEFVRFDSVRVNPKPVRDRRVPVMVGGNS
MUL_3448|M.ulcerans_Agy99 EYVAAMRTLWRD---DLASFTGEFVRFDSVRVNPKPVRDRRVPVMVGGNS
MSMEG_2811|M.smegmatis_MC2_155 EYVEAMRTLWRQ---DVADFTGEFAAFDAVRLHPKPSAG-SIPIIVGGNG
Mflv_4839|M.gilvum_PYR-GCK EYVGAMRTLWRD---DDASFAGRYVAFDGVRANPKPQGR-SIPVVLGGNS
Mvan_5752|M.vanbaalenii_PYR-1 EQVELLRQLWTQR---TVTFDGRHHRVTGAGIAPLPVQR-PIPLWIGAAA
TH_2945|M.thermoresistible__bu EYLAAIIELWTQ---DEPSFAGQFVSFEDVVFEPKPIQQPHPPIWIGGDS
MLBr_00131|M.leprae_Br4923 EALATIRALWDSN-GELVSRESQFFPLHNALFDLPPYRGKWPEIWVAAHG
* : : :* . . . * : :.. .
Mb3106c|M.bovis_AF2122/97 PANFPRIARLNAGWIAISPSP----QRLSGPLQRLRAMAGGDVP------
Rv3079c|M.tuberculosis_H37Rv PANFPRIARLNAGWIAISPSP----QRLSGPLQRLRAMAGGDVP------
MAB_0366|M.abscessus_ATCC_1997 KAAARRAGRLGSGFQPLGLVG-DDLTAAVGVMRAAAVDAGRDPRGL----
MAV_2935|M.avium_104 RAAARRAGRFGDGFQPLGVTG-ARLASLIGLMRDEAVAAGRDPAAL----
MMAR_1586|M.marinum_M DAALRRVAAWADGWYGFNLDGVGAVRERVGTLDRLCAESGRNRD------
MUL_3448|M.ulcerans_Agy99 DAALRRVAAWADGWYGFNLDGVGAVRERVGTLDRLCAESGRNRD------
MSMEG_2811|M.smegmatis_MC2_155 DRALARVASWGDGWYGFNLDGVAAVGERMDMLRRLCARAGRDVD------
Mflv_4839|M.gilvum_PYR-GCK DAALRRVAEWGDGWYGFDLADVEEAAFRASKLRSMCREFGRDPT------
Mvan_5752|M.vanbaalenii_PYR-1 PRGYARAGRIADGWFPLVGPG-PALDEAKGAVDAAAREAGRDPSEIG---
TH_2945|M.thermoresistible__bu DAALRRAARFASGWFTFLTKP-EDIAARVDIITSQPEYDGRPFDVMHGLG
MLBr_00131|M.leprae_Br4923 PRMLQATGRYADAWIPIVLVRPTDYSCALEVVRTAASDAGRDPMSITPAA
. .: : : *
Mb3106c|M.bovis_AF2122/97 ----------------------------VTVCQWGEAAAKDLEGYRHLGV
Rv3079c|M.tuberculosis_H37Rv ----------------------------VTVCQWGEAAAKDLEGYRHLGV
MAB_0366|M.abscessus_ATCC_1997 --------------------------DLVLGHGLHRVDQAALDQAAHAGA
MAV_2935|M.avium_104 --------------------------EVSLGHSVSKIDAERAARLADQGA
MMAR_1586|M.marinum_M --------------------------ELRLSVALRCPQPCDVDALAELGI
MUL_3448|M.ulcerans_Agy99 --------------------------ELRLSVALRCPQPCDVDALAELGI
MSMEG_2811|M.smegmatis_MC2_155 --------------------------DLHCAVALQAPAVGDIPALVDLGV
Mflv_4839|M.gilvum_PYR-GCK --------------------------ELKLSVALHDPVPADAVRLADAGI
Mvan_5752|M.vanbaalenii_PYR-1 -----------------------MEGRVRWRHDLDQTAADIGEWAEHGAS
TH_2945|M.thermoresistible__bu TNRVGE--------------GHVPTGDRGAWPGMSAAELIDRIGALHEHG
MLBr_00131|M.leprae_Br4923 VRGIITGRTRDDVDEALDSVLVRMIALGVPGEAWARHGVEHPMGADFAGV
Mb3106c|M.bovis_AF2122/97 ER-VLLELPTEPRDPTLRYLDKLQAELARLA-------------------
Rv3079c|M.tuberculosis_H37Rv ER-VLLELPTEPRDPTLRYLDKLQAELARLA-------------------
MAB_0366|M.abscessus_ATCC_1997 GRMLLSASRRATTLEAVLEEMAGCATRLELG---GPR-------------
MAV_2935|M.avium_104 DRLVLAMPP-TSDIEQAKDMLSACAQRLSLRPVSGP--------------
MMAR_1586|M.marinum_M DELVLVETPPEAADAAQNWVSALAERWLSTVWVR----------------
MUL_3448|M.ulcerans_Agy99 DELVLVETPPEAADAAQNWVSALAERWLSTVWVR----------------
MSMEG_2811|M.smegmatis_MC2_155 DELVIVESPPQDPDVAADWVDALADRWITCI-------------------
Mflv_4839|M.gilvum_PYR-GCK DELVLVAGPPADPIAAEAWIADLAGQWMRVRT------------------
Mvan_5752|M.vanbaalenii_PYR-1 HLSVDTMRAGLRTVNEHLAVLEAIADRVGLR-------------------
TH_2945|M.thermoresistible__bu VTYTSVPIPPVRGVEEYLDYSQWVMEEIAPNVP-----------------
MLBr_00131|M.leprae_Br4923 QDIIPQTIDEETVVSYAAKVPAALMKEVLFSGTPEEVIDQVAEWRDHGLK
Mb3106c|M.bovis_AF2122/97 ----------------------------------
Rv3079c|M.tuberculosis_H37Rv ----------------------------------
MAB_0366|M.abscessus_ATCC_1997 ----------------------------------
MAV_2935|M.avium_104 ----------------------------------
MMAR_1586|M.marinum_M ----------------------------------
MUL_3448|M.ulcerans_Agy99 ----------------------------------
MSMEG_2811|M.smegmatis_MC2_155 ----------------------------------
Mflv_4839|M.gilvum_PYR-GCK ----------------------------------
Mvan_5752|M.vanbaalenii_PYR-1 ----------------------------------
TH_2945|M.thermoresistible__bu ----------------------------------
MLBr_00131|M.leprae_Br4923 YLVVINGSLVNSSLRKTVSALLPHARVLRGLKKL