For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGRKPQYSADDILDAALGLVAAGGPSAATATAIGRVLGAPSGSIYHRFPSRDELMARLWLRSIARYQDGV LTALALPDADDALAAAIDHAFDWTASNHNEALLLLQYEKADLVAHWPDTLATELKSLNSRVRKTLREYTD RRYGKVTTERIDKVMFALIDMPYAAVRRHLPDKGEPAPWLREFVHTSAHALLARACLNEP
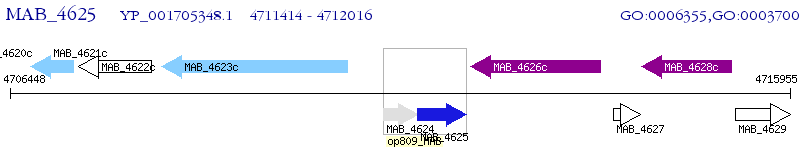
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4625 | - | - | 100% (200) | TetR family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_3395 | - | 1e-22 | 33.51% (191) | putative transcriptional regulator, TetR |
| M. abscessus ATCC 19977 | MAB_2466c | - | 2e-10 | 23.92% (209) | putative transcriptional regulator, TetR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4881 | - | 1e-26 | 37.17% (191) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_02497 | - | 4e-05 | 25.49% (153) | hypothetical protein MLBr_02497 |
| M. marinum M | MMAR_1145 | - | 8e-09 | 30.00% (120) | transcriptional regulatory protein |
| M. avium 104 | MAV_4715 | - | 2e-25 | 37.36% (182) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_0859 | - | 4e-26 | 37.57% (189) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_2354 | - | 3e-06 | 27.34% (128) | putative transcriptional regulator |
| M. ulcerans Agy99 | MUL_0910 | - | 2e-08 | 29.17% (120) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_1548 | - | 3e-25 | 35.75% (193) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1145|M.marinum_M ------------------MAATRTTARAGGDRRAAIIEAALNRFLSQGID
MUL_0910|M.ulcerans_Agy99 ------------------MAATRTTARAGGDRRAAIIEAALNRFLSQGID
Mflv_4881|M.gilvum_PYR-GCK ------------------------MPRPARYTVDVLLDAAAELLAAEGPA
Mvan_1548|M.vanbaalenii_PYR-1 ------------------------MPRPARYTTDVLLDAAAELLAAEGPA
MAB_4625|M.abscessus_ATCC_1997 ------------------------MGRKPQYSADDILDAALGLVAAGGPS
MAV_4715|M.avium_104 ----------------------------------MILDAARALVLDGGPR
MSMEG_0859|M.smegmatis_MC2_155 ------------------------MAPPRKHETDAILDAARALVLTEGPR
MLBr_02497|M.leprae_Br4923 --------------------------------------------------
TH_2354|M.thermoresistible__bu MPGVSVLKSPGGPPGARPAEVPRRANRRGQATRENMLEAALAALATGDPG
MMAR_1145|M.marinum_M ATALRQIQRDAGVSNGSFFHHFPSKEVLAAAVYVDCVTRYQQALLKDLAR
MUL_0910|M.ulcerans_Agy99 ATALRQIQRDAGVSNGSFFHHFPSKEVLTAAVYVDCVTRYQQALLKDLVR
Mflv_4881|M.gilvum_PYR-GCK AVTMSAVARSTGAPSGSVYHRFPTRAALCGELWVRTEERFQEELLEALSG
Mvan_1548|M.vanbaalenii_PYR-1 AVTMSAVARSTGAPSGSVYHRFPTRAALCGELWVRTEERFQGELIEALSV
MAB_4625|M.abscessus_ATCC_1997 AATATAIGRVLGAPSGSIYHRFPSRDELMARLWLRSIARYQDGVLTALAL
MAV_4715|M.avium_104 TASVAAIATASGAPAGTLYHRFGNRDGILTAAWLRALERFQARALAADGD
MSMEG_0859|M.smegmatis_MC2_155 AAGVAAIAKASGAPVGTLYHRFGNRDGILKATWLRGIERFQRRAMAADGA
MLBr_02497|M.leprae_Br4923 ------MFASRAALLGRVWLRAGYR-------FLKTQTALVDQALSASVR
TH_2354|M.thermoresistible__bu AVSANRIAKQIGVTWGAVKYQFGDIDGLWAAVLRRTAER-RAGVIGRHDA
: .. * . : :
MMAR_1145|M.marinum_M YPDAESAVRGIVGMHLSWCTEHPEMARFLITMTEPAVHRAAADE-LRSHN
MUL_0910|M.ulcerans_Agy99 YPDAESAVRGIVGMHLSWCTEHPEMARFLITMTEPAVHRAAADE-LRSHN
Mflv_4881|M.gilvum_PYR-GCK DGDPQNRCVAGALRNMQWCRENPVEAHVLLAGPDQLDMGEWPDT-VAGRR
Mvan_1548|M.vanbaalenii_PYR-1 DGDAQQRCVSGALRIVQWCRDNPVEAQVMLAGPTELGMGDWPEP-VTSRR
MAB_4625|M.abscessus_ATCC_1997 P-DADDALAAAIDHAFDWTASNHNEALLLLQYEKADLVAHWPDT-LATEL
MAV_4715|M.avium_104 T--PEDTAVAMAVAAVSFARALPDDARLLLTIRPGDLLDGEPDAAFRQTL
MSMEG_0859|M.smegmatis_MC2_155 T--PVDVAVAMAGAVLQFARDEPEDARLLLSIRRADLLDGGPDADFDATL
MLBr_02497|M.leprae_Br4923 R-AGVEAVVAAADAPVVFAEQYPDSSRLLLTVRREELLKSFLPNEIEIEL
TH_2354|M.thermoresistible__bu GAPLRERVAGIIDMLYDGLTAS--DSRAIENLRAALPRDRAELERLYPRT
. . : : .
MMAR_1145|M.marinum_M ERFATAVQTWWRPHAH--YGALRP--LSPAHSQALWLGPAQELVRAWLLG
MUL_0910|M.ulcerans_Agy99 ERFATAVQTWWRPHAH--YGALRP--LSPAHSQALWLGPAQELVRAWLLG
Mflv_4881|M.gilvum_PYR-GCK KRLQRKLRKVLAEIAG-DHGRIDG--RVDGRVAAAVMDVPAAIVRRHLRA
Mvan_1548|M.vanbaalenii_PYR-1 KRLQRKLRKALSDLSD-DTARVN----------AAIVDVPAAIVRRHLRA
MAB_4625|M.abscessus_ATCC_1997 KSLNSRVRKTLREYTDRRYGKVTT--ERIDKVMFALIDMPYAAVRRHLPD
MAV_4715|M.avium_104 AAMNAPLTQRLAQLARHLYGNARP--RSVDAVARAVADLPYAVVRRHAHD
MSMEG_0859|M.smegmatis_MC2_155 ADMNAPVFERLHELARGIFGRDDD--RAMDAVMRAVADLPYATVRRHIDD
MLBr_02497|M.leprae_Br4923 RELDALLISLLTRLAVVIWDRKDT--GAIDTITTCLMDLPTAVLLHRDRL
TH_2354|M.thermoresistible__bu AAELSSWGEGWLQTCQQAFADLDVDPERVAEVASFIPGAMRGITSERQLG
.
MMAR_1145|M.marinum_M NLAAPPNAADAAILADAAWLCLRAGPAG----------------------
MUL_0910|M.ulcerans_Agy99 NLAAPPNAADAAILADAAWLCLRAGPAG----------------------
Mflv_4881|M.gilvum_PYR-GCK RQSIPAAADDIVADCARALICPN---------------------------
Mvan_1548|M.vanbaalenii_PYR-1 GQTIPAVADAIVEECARALIAPS---------------------------
MAB_4625|M.abscessus_ATCC_1997 KGEPAPWLREFVHTSAHALLARACLNEP----------------------
MAV_4715|M.avium_104 -DPMPSWLETDVAASARAVLRS----------------------------
MSMEG_0859|M.smegmatis_MC2_155 -GKWPVWLSEDLATSVRRLLSVDKIVSASREIAAPSDVIFELIADPAQQP
MLBr_02497|M.leprae_Br4923 GNKTAR---KHLHAAVRAVLEVGPPP------------------------
TH_2354|M.thermoresistible__bu TYIDLELARRGLTNAIVSYLEDRGPGEKSNQ-------------------
. :
MMAR_1145|M.marinum_M --------------------------------------------------
MUL_0910|M.ulcerans_Agy99 --------------------------------------------------
Mflv_4881|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1548|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4625|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_4715|M.avium_104 --------------------------------------------------
MSMEG_0859|M.smegmatis_MC2_155 RWDGNENLLRMASGGRVRGVGEVFTMEIKQGGFRDNHVIEFEEGRRIAWR
MLBr_02497|M.leprae_Br4923 --------------------------------------------------
TH_2354|M.thermoresistible__bu --------------------------------------------------
MMAR_1145|M.marinum_M --------------------------------------------------
MUL_0910|M.ulcerans_Agy99 --------------------------------------------------
Mflv_4881|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1548|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4625|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_4715|M.avium_104 ---FGERP------------------------------------------
MSMEG_0859|M.smegmatis_MC2_155 PAVAGEQPLGHLWRWELQPAGEGRTLVTHTYDWTQLTDEKRFPKASATTA
MLBr_02497|M.leprae_Br4923 --------------------------------------------------
TH_2354|M.thermoresistible__bu --------------------------------------------------
MMAR_1145|M.marinum_M ----------------
MUL_0910|M.ulcerans_Agy99 ----------------
Mflv_4881|M.gilvum_PYR-GCK ----------------
Mvan_1548|M.vanbaalenii_PYR-1 ----------------
MAB_4625|M.abscessus_ATCC_1997 ----------------
MAV_4715|M.avium_104 ----------------
MSMEG_0859|M.smegmatis_MC2_155 DKLQESVDRLAAIVEG
MLBr_02497|M.leprae_Br4923 ----------------
TH_2354|M.thermoresistible__bu ----------------