For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAPERKHASDAILDAARSLLLDGGPRAATIAAIAGASGAPPGTLNHRFGNRAAIMSATWLRAVERFHEHA LRALHSAGDPVETAVALSLSVPAFARAYPEDARLLLSLRSADILGAEANSTLDEMNAPLFTAMRDLTRAI YQRGDARSRDRLLRAVVDLPYAAVRRHMPTLPGWLEEDLAAAVRALLASR
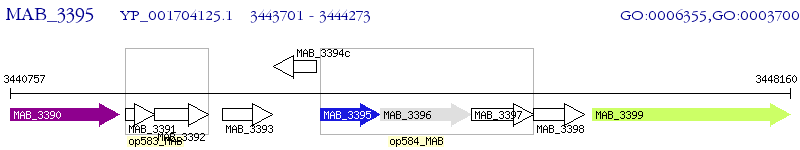
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3395 | - | - | 100% (190) | putative transcriptional regulator, TetR |
| M. abscessus ATCC 19977 | MAB_4625 | - | 1e-22 | 33.51% (191) | TetR family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_2466c | - | 2e-19 | 32.31% (195) | putative transcriptional regulator, TetR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3081 | - | 3e-05 | 28.31% (166) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_4881 | - | 4e-22 | 35.64% (188) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3055 | - | 3e-05 | 28.31% (166) | TetR family transcriptional regulator |
| M. leprae Br4923 | MLBr_02497 | - | 5e-11 | 32.89% (152) | hypothetical protein MLBr_02497 |
| M. marinum M | MMAR_4335 | - | 7e-07 | 30.69% (101) | TetR family transcriptional regulator |
| M. avium 104 | MAV_4715 | - | 6e-47 | 54.35% (184) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_0859 | - | 4e-52 | 56.19% (194) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_2712 | - | 2e-06 | 46.43% (56) | PUTATIVE transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_0145 | - | 5e-07 | 30.69% (101) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1548 | - | 5e-20 | 31.38% (188) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_4715|M.avium_104 --------------------------------------MILDAARALVLD
MSMEG_0859|M.smegmatis_MC2_155 ----------------------------MAPPRKHETDAILDAARALVLT
MAB_3395|M.abscessus_ATCC_1997 ----------------------------MAPERKHASDAILDAARSLLLD
MMAR_4335|M.marinum_M ---MSTDSHG-----------------RRWAKTDATQRRILDAATEVFAT
MUL_0145|M.ulcerans_Agy99 ---MSTDSHG-----------------RRWAKTDATQRRILDAATEVFAT
Mb3081|M.bovis_AF2122/97 ---MSGAERLGDLPVFARQEP-----VPERGDAARNRALLLEAARRLIAR
Rv3055|M.tuberculosis_H37Rv ---MSGAERLGDLPVFARQEP-----VPERGDAARNRALLLEAARRLIAR
TH_2712|M.thermoresistible__bu VSLMSADELLPARPATDVAAPTATTAPPEREDAARNRALLLDAARRLIAE
Mflv_4881|M.gilvum_PYR-GCK ----------------------------MPRPARYTVDVLLDAAAELLAA
Mvan_1548|M.vanbaalenii_PYR-1 ----------------------------MPRPARYTTDVLLDAAAELLAA
MLBr_02497|M.leprae_Br4923 --------------------------------------------------
MAV_4715|M.avium_104 GGPRTASVAAIATASGAPAGTLYHRFGNRDGILTAAWLRALERFQARALA
MSMEG_0859|M.smegmatis_MC2_155 EGPRAAGVAAIAKASGAPVGTLYHRFGNRDGILKATWLRGIERFQRRAMA
MAB_3395|M.abscessus_ATCC_1997 GGPRAATIAAIAGASGAPPGTLNHRFGNRAAIMSATWLRAVERFHEHALR
MMAR_4335|M.marinum_M RGFNAATMAEVVASSGASIGSIYHHFGGKTELFLAIFEQMASAVEGKIEE
MUL_0145|M.ulcerans_Agy99 RGFNAATMAEVVASSGASIGSIYHHFGGKTELFLAIFEQMASAVEGKIEE
Mb3081|M.bovis_AF2122/97 SGADAITMDDVAAAAGVGKGTLFRRFGSRAGLMMVLLDEDERASQQAFLF
Rv3055|M.tuberculosis_H37Rv SGADAITMDDVAAAAGVGKGTLFRRFGSRAGLMMVLLDEDERASQQAFLF
TH_2712|M.thermoresistible__bu RGAEAVTMDDIAAAAGVGKGTLFRRFGSRAGLMLELLHDDETAFQQAFMF
Mflv_4881|M.gilvum_PYR-GCK EGPAAVTMSAVARSTGAPSGSVYHRFPTRAALCGELWVRTEERFQEELLE
Mvan_1548|M.vanbaalenii_PYR-1 EGPAAVTMSAVARSTGAPSGSVYHRFPTRAALCGELWVRTEERFQGELIE
MLBr_02497|M.leprae_Br4923 ----------MFASRAALLGRVWLRAGYR-------FLKTQTALVDQALS
: : .. * : : :
MAV_4715|M.avium_104 ADGDT-----PEDTAVAMAVAAVSFARALPDDARLLLTIRPGDLLDGEPD
MSMEG_0859|M.smegmatis_MC2_155 ADGAT-----PVDVAVAMAGAVLQFARDEPEDARLLLSIRRADLLDGGPD
MAB_3395|M.abscessus_ATCC_1997 ALHSAG---DPVETAVALSLSVPAFARAYPEDARLLLSLRSADILG----
MMAR_4335|M.marinum_M ALQQAG-DETDRRRIFELGVRAYLDGLWEHRRVARLLAS------GDRPT
MUL_0145|M.ulcerans_Agy99 ALQQAG-DETDRRRIFELGVRAYLDGLWEHRRVARLLAS------GDRPT
Mb3081|M.bovis_AF2122/97 GPPPLGPDAPPLDRLIAFGRERMR---FVHAHHQLLSEA------NRDPQ
Rv3055|M.tuberculosis_H37Rv GPPPLGPDAPPLDRLIAFGRERMR---FVHAHHQLLSEA------NRDPQ
TH_2712|M.thermoresistible__bu GPPPLGPGAEPLERLLAYGRRRLQ---FVCDHHALLTDV------SREPQ
Mflv_4881|M.gilvum_PYR-GCK ALSGDG---DPQNRCVAGALRNMQWCRENPVEAHVLLAGPDQLDMGEWPD
Mvan_1548|M.vanbaalenii_PYR-1 ALSVDG---DAQQRCVSGALRIVQWCRDNPVEAQVMLAGPTELGMGDWPE
MLBr_02497|M.leprae_Br4923 ASVRRA----GVEAVVAAADAPVVFAEQYPDSSRLLLTVRREELLKSFLP
. . . :
MAV_4715|M.avium_104 AAFRQTLAAMNAPLTQRLAQLARHLYGNARPRSVDAVARAVADLPYAVVR
MSMEG_0859|M.smegmatis_MC2_155 ADFDATLADMNAPVFERLHELARGIFGRDDDRAMDAVMRAVADLPYATVR
MAB_3395|M.abscessus_ATCC_1997 AEANSTLDEMNAPLFTAMRDLTRAIYQRGDARSRDRLLRAVVDLPYAAVR
MMAR_4335|M.marinum_M DFGITRRDRMTSAFRSWMAVLE--LDSSLHGQ--FLSRALLAIMEESSLM
MUL_0145|M.ulcerans_Agy99 DFGITRRDRMTSAFRSWMAVLE--LDSSLHGQ--FLSRALLR--------
Mb3081|M.bovis_AF2122/97 TRHSAALSVLRTHLRVLLASAP--TTGDLD----AQTDALLALLDVDYVE
Rv3055|M.tuberculosis_H37Rv TRHSAALSVLRTHLRVLLASAP--TTGDLD----AQTDALLALLDVDYVE
TH_2712|M.thermoresistible__bu TRFGPPMTLHHTHIRSLLAAAD--ADGDLD----VQADALLALTDPDYVR
Mflv_4881|M.gilvum_PYR-GCK TVAGRRKRLQRKLRKVLAEIAG--DHGRIDGRVDGRVAAAVMDVPAAIVR
Mvan_1548|M.vanbaalenii_PYR-1 PVTSRRKRLQRKLRKALSDLSD--DTARVN--------AAIVDVPAAIVR
MLBr_02497|M.leprae_Br4923 NEIEIELRELDALLISLLTRLAVVIWDRKDTGAIDTITTCLMDLPTAVLL
:
MAV_4715|M.avium_104 RHAHDDPMPSWLETDVAASARAVLRS------------------------
MSMEG_0859|M.smegmatis_MC2_155 RHIDDGKWPVWLSEDLATSVRRLLSVDKIVSASREIAAPSDVIFELIADP
MAB_3395|M.abscessus_ATCC_1997 RHMP--TLPGWLEEDLAAAVRALLASR-----------------------
MMAR_4335|M.marinum_M VAACEEPEQVKPVIDATIEWIGRLTE------------------------
MUL_0145|M.ulcerans_Agy99 ------------------SWRSRH--------------------------
Mb3081|M.bovis_AF2122/97 HQLNAGGHTLQTLGDAWESLARKLCGR-----------------------
Rv3055|M.tuberculosis_H37Rv HQLNAGGHTLQTLGDAWESLARKLCGR-----------------------
TH_2712|M.thermoresistible__bu HQVTDRGVALDALADSWGDVARKLCRR-----------------------
Mflv_4881|M.gilvum_PYR-GCK RHLRARQSIPAAADDIVADCARALICPN----------------------
Mvan_1548|M.vanbaalenii_PYR-1 RHLRAGQTIPAVADAIVEECARALIAPS----------------------
MLBr_02497|M.leprae_Br4923 HRDRLGNKTARKHLHAAVRAVLEVGPPP----------------------
MAV_4715|M.avium_104 --------------------------------------------------
MSMEG_0859|M.smegmatis_MC2_155 AQQPRWDGNENLLRMASGGRVRGVGEVFTMEIKQGGFRDNHVIEFEEGRR
MAB_3395|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_4335|M.marinum_M --------------------------------------------------
MUL_0145|M.ulcerans_Agy99 --------------------------------------------------
Mb3081|M.bovis_AF2122/97 --------------------------------------------------
Rv3055|M.tuberculosis_H37Rv --------------------------------------------------
TH_2712|M.thermoresistible__bu --------------------------------------------------
Mflv_4881|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1548|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_02497|M.leprae_Br4923 --------------------------------------------------
MAV_4715|M.avium_104 -------FGERP--------------------------------------
MSMEG_0859|M.smegmatis_MC2_155 IAWRPAVAGEQPLGHLWRWELQPAGEGRTLVTHTYDWTQLTDEKRFPKAS
MAB_3395|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_4335|M.marinum_M --------------------------------------------------
MUL_0145|M.ulcerans_Agy99 --------------------------------------------------
Mb3081|M.bovis_AF2122/97 --------------------------------------------------
Rv3055|M.tuberculosis_H37Rv --------------------------------------------------
TH_2712|M.thermoresistible__bu --------------------------------------------------
Mflv_4881|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1548|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_02497|M.leprae_Br4923 --------------------------------------------------
MAV_4715|M.avium_104 --------------------
MSMEG_0859|M.smegmatis_MC2_155 ATTADKLQESVDRLAAIVEG
MAB_3395|M.abscessus_ATCC_1997 --------------------
MMAR_4335|M.marinum_M --------------------
MUL_0145|M.ulcerans_Agy99 --------------------
Mb3081|M.bovis_AF2122/97 --------------------
Rv3055|M.tuberculosis_H37Rv --------------------
TH_2712|M.thermoresistible__bu --------------------
Mflv_4881|M.gilvum_PYR-GCK --------------------
Mvan_1548|M.vanbaalenii_PYR-1 --------------------
MLBr_02497|M.leprae_Br4923 --------------------