For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGTAVQLAGKVVAVTGGARGIGRAIATAFAAEGAKVAIGDIDKKLCENTAAEIGNSAIGLPLDVTDHGSF EAFFDTIAATVGPVDVIVNNAGIMPITPFDDESLESIQRQLDINVRGVMWGSQLAIKRMKPRGGGVIVNI ASAAGKAGFPGLATYCATKHAVVGLSESLSLEYEPAGISVVCVMPGMVNTELISGLDTHWLLKTVEPEDI ANAVVKAVRRGVFPVFVPKMFGGMYRTMSVLPRSTGKFLTRKLGIQDFMLNAHGSDARKAYEDRASHSGP STE
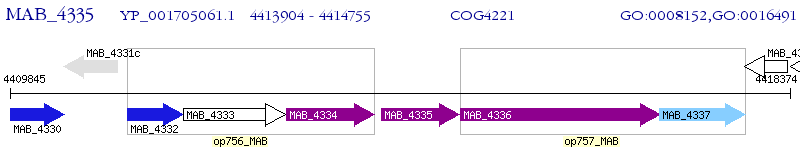
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4335 | - | - | 100% (283) | putative short-chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_3405 | - | 6e-55 | 44.93% (276) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1526 | - | 2e-49 | 41.48% (270) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1896c | - | 3e-56 | 45.86% (266) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0217 | - | 8e-53 | 42.65% (272) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv1865c | - | 3e-56 | 45.86% (266) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00862 | ephD | 5e-31 | 32.58% (264) | short chain dehydrogenase |
| M. marinum M | MMAR_2743 | - | 6e-57 | 45.76% (271) | short-chain type dehydrogenase |
| M. avium 104 | MAV_2700 | - | 4e-64 | 49.82% (275) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0771 | - | 7e-53 | 43.01% (272) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3427 | - | 2e-49 | 41.82% (275) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3011 | - | 2e-56 | 45.39% (271) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_2345 | - | 3e-66 | 51.12% (268) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_2345|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2700|M.avium_104 --------------------------------------------------
Mb1896c|M.bovis_AF2122/97 --------------------------------------------------
Rv1865c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2743|M.marinum_M --------------------------------------------------
MUL_3011|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0217|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0771|M.smegmatis_MC2_155 --------------------------------------------------
TH_3427|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIVLVHGFPDSHVLWD
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_2345|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2700|M.avium_104 --------------------------------------------------
Mb1896c|M.bovis_AF2122/97 --------------------------------------------------
Rv1865c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2743|M.marinum_M --------------------------------------------------
MUL_3011|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0217|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0771|M.smegmatis_MC2_155 --------------------------------------------------
TH_3427|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVVPLLAKRFRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDELSP
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_2345|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2700|M.avium_104 --------------------------------------------------
Mb1896c|M.bovis_AF2122/97 --------------------------------------------------
Rv1865c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2743|M.marinum_M --------------------------------------------------
MUL_3011|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0217|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0771|M.smegmatis_MC2_155 --------------------------------------------------
TH_3427|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSG
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_2345|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2700|M.avium_104 --------------------------------------------------
Mb1896c|M.bovis_AF2122/97 --------------------------------------------------
Rv1865c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2743|M.marinum_M --------------------------------------------------
MUL_3011|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0217|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0771|M.smegmatis_MC2_155 --------------------------------------------------
TH_3427|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMV
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_2345|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2700|M.avium_104 --------------------------------------------------
Mb1896c|M.bovis_AF2122/97 --------------------------------------------------
Rv1865c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2743|M.marinum_M --------------------------------------------------
MUL_3011|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0217|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0771|M.smegmatis_MC2_155 --------------------------------------------------
TH_3427|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQ
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_2345|M.vanbaalenii_PYR-1 ------------------------------------MVPSRPARGPRTSA
MAV_2700|M.avium_104 --------------------------------------------------
Mb1896c|M.bovis_AF2122/97 --------------------------------------------------
Rv1865c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2743|M.marinum_M --------------------------------------------------
MUL_3011|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0217|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0771|M.smegmatis_MC2_155 --------------------------------------------------
TH_3427|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAAVHD
MAB_4335|M.abscessus_ATCC_1997 --------MGTAVQLA-----------GKVVAVTGGARGIGRAIATAFAA
Mvan_2345|M.vanbaalenii_PYR-1 QLGMKGGTVTADRNLT-----------GRVVAITGAARGIGAATARAFAA
MAV_2700|M.avium_104 ------------MDLT-----------DKVVAITGGARGIGLATAKAVLA
Mb1896c|M.bovis_AF2122/97 ----MPGRTSIGVKIRDKVQ-------DKVIAITGGARGIGLATAAALHN
Rv1865c|M.tuberculosis_H37Rv ----MPGRTSIGVKIRDKVQ-------DKVIAITGGARGIGLATAAALHN
MMAR_2743|M.marinum_M ----MADSTTIGVRVR-----------DKVIVITGGARGIGLATATALHK
MUL_3011|M.ulcerans_Agy99 ----MADSTTIGVRVR-----------NKVIVITGGARGIGLATATALHK
Mflv_0217|M.gilvum_PYR-GCK ----MDN-------IR-----------GKTIAITGAARGIGYATAEALLA
MSMEG_0771|M.smegmatis_MC2_155 ----MDN-------IR-----------GKTIAITGAARGIGYATAKALLA
TH_3427|M.thermoresistible__bu ----MDN-------IR-----------GKTIAITGAARGIGYATATALLA
MLBr_00862|M.leprae_Br4923 LADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAGSGIGRETALALAR
: . : :**.. *** * *.
MAB_4335|M.abscessus_ATCC_1997 EGAKVAIGDIDKKLCENTAAEIGN---SAIGLPLDVTDHGSFEAFFDTIA
Mvan_2345|M.vanbaalenii_PYR-1 AGASVAIGDLDADLAVRTAAEIG-----CLGSGVDVTDHDSVEKFLDEVE
MAV_2700|M.avium_104 EGAKVALGDLDTELAEKQAVELGG-DPAVVGLPLDVSDPASFAAFLDDVE
Mb1896c|M.bovis_AF2122/97 LGAKVAIGDIDEAMAKESGADLD--LDMYG--KLDVTDPDSFSGFLDAVE
Rv1865c|M.tuberculosis_H37Rv LGAKVAIGDIDEAMAKESGADLD--LDMYG--KLDVTDPDSFSGFLDAVE
MMAR_2743|M.marinum_M LGAKVAIGDVDEPAVKEAGADLG--LEVYG--KLDVTDPNSFSDFLDQVE
MUL_3011|M.ulcerans_Agy99 LGAKVAIGDVGEPAVKEAGADLG--LEVYG--KLDVTDPNSFSDFLDQVE
Mflv_0217|M.gilvum_PYR-GCK HGARVVIGDRDVALQESSVAKLTN-LGPVSGYPLDVTDQESFATFLDKAR
MSMEG_0771|M.smegmatis_MC2_155 HGARVVIGDRDVALQESAVVELSK-LGQVSGYPLDVTDRESFATFLDKAR
TH_3427|M.thermoresistible__bu RGARVVIGDRDVALQESAVAQLGR-LGPVSGYPLDVTDRESFATFLDKAR
MLBr_00862|M.leprae_Br4923 RGAEVVVSDIDEAAAKNTAAQIVASGGVAYAYALDVSDAAAVEAFAEQVS
** *.:.* . ..: :**:* :. * :
MAB_4335|M.abscessus_ATCC_1997 ATVG-PVDVIVNNAGIMPITPFDDESLESIQRQLDINVRGVMWGSQLAIK
Mvan_2345|M.vanbaalenii_PYR-1 QRLG-PLDVMVNNAGIMPVTQMLDESPESVRRQLAVNVAGVVFGTQHAAS
MAV_2700|M.avium_104 ARLG-RLDVLVNNAGIMPTGPFVDESPTMSRRMIDVNVYGVLNGSRLAAA
Mb1896c|M.bovis_AF2122/97 RQLG-PIDVLVNNAGIMPVGRIVDEPDPVTRRILDINVYGVILGSKLAAQ
Rv1865c|M.tuberculosis_H37Rv RQLG-PIDVLVNNAGIMPVGRIVDEPDPVTRRILDINVYGVILGSKLAAQ
MMAR_2743|M.marinum_M RQLG-PLDVLVNNAGIMPVGRIVDEPDSVTRRILDINVYGVMVGSKLAAQ
MUL_3011|M.ulcerans_Agy99 RQLG-PLDVLVNNAGIMPVGRIVDEPDSVTRRILDINVYGVMVGSKLAAQ
Mflv_0217|M.gilvum_PYR-GCK TDGGGHIDVLINNAGVMPVGPFLDETEQSIRSSLEVNVYGVLTGCRLALP
MSMEG_0771|M.smegmatis_MC2_155 TDGGGHIDVLINNAGVMPVGPFLEQSEQSIRSNIEVNVYGVLTGCQLALP
TH_3427|M.thermoresistible__bu TDGGGHIDVLINNAGVMPIGPFLEQSEQAIRSSIEVNLYGVLTGCQLALP
MLBr_00862|M.leprae_Br4923 AKHG-VPDIVVNNAGIGQAGRFLDTPPEEFDRVLAVNLGGVVNCCRAFGQ
* *:::****: : : . : :*: **: :
MAB_4335|M.abscessus_ATCC_1997 RMKPRGGG-VIVNIASAAGKAGFPGLATYCATKHAVVGLSESLSLEYEPA
Mvan_2345|M.vanbaalenii_PYR-1 RMVARRSG-HIVNVASAAGRIAFGGVATYTATKFAVVGFTEAAALELRDS
MAV_2700|M.avium_104 RFVPRGAG-HIVNIASLAGVTGEPGMATYCGTKHFVVGFTESLHRELRPH
Mb1896c|M.bovis_AF2122/97 RMVPRGRG-HVINVASLAGEIYAVGVATYCASKHAVVAFTDSARLEYRSA
Rv1865c|M.tuberculosis_H37Rv RMVPRGRG-HVINVASLAGEIYAVGVATYCASKHAVVAFTDSARLEYRSA
MMAR_2743|M.marinum_M RMVPRGRG-HVINVASLAGEIYVVGLATYCASKHAVIAFTDAARIEYRST
MUL_3011|M.ulcerans_Agy99 RMVPRGRG-HVINVASLAGEIYVVGLATYCASKHAVIAFTDAARIEYRTT
Mflv_0217|M.gilvum_PYR-GCK DMVKRRSG-HIINIASLSGVIPLPGQVVYVGAKYAVVGLTTALADEMAPH
MSMEG_0771|M.smegmatis_MC2_155 DMVKRRRG-HIINIASLSGVIPLPGQVVYVGAKYAVVGLSTALADEMAPY
TH_3427|M.thermoresistible__bu DMVRRRSG-HIINIASMSGLIPVPGQVPYNAAKFGVVGLSVALADEMAPY
MLBr_00862|M.leprae_Br4923 RLVERGTGGHIVNVSSMAAYAPLQSLSAYCTSKAATYMFSDCLRAELDAV
: * * ::*::* :. . * :* . :: . *
MAB_4335|M.abscessus_ATCC_1997 GISVVCVMPGMVNTELISGLDTH---WLLKTVEPEDIANAVVKAVRRGVF
Mvan_2345|M.vanbaalenii_PYR-1 GVRFTCVLPGVVNTELTSGLRDH---WLLRSCEPEEVAAGVVAGVRRHRR
MAV_2700|M.avium_104 RVGVSLVLPGIINTELSAGTKVPGWARPLATAEPEDVAAGIVAAVSKDKP
Mb1896c|M.bovis_AF2122/97 GVKFSMVLPSFVNTELIAGTGGI---KGFKNAEPADIADAIVGLIVHPKP
Rv1865c|M.tuberculosis_H37Rv GVKFSMVLPSFVNTELIAGTGGI---KGFKNAEPADIADAIVGLIVHPKP
MMAR_2743|M.marinum_M GVKFSMVLPTFVNTELASGTPGM---KGFKNAEPSDIADAIVALVANPKP
MUL_3011|M.ulcerans_Agy99 GVKFSMVLPTFVNTELASGTPGM---KGFKNAEPSDIADAIVALVANPKP
Mflv_0217|M.gilvum_PYR-GCK GVNVSVIMPPFTNTELIAGTKSG---GAIKPVEPEDIAAAIVKTLNKPKT
MSMEG_0771|M.smegmatis_MC2_155 GVDVSVIMPPFTNTDLISGTKSG---GAIKPVEPEEIAAAIVKTLNKPKT
TH_3427|M.thermoresistible__bu GVEVSVVMPPFTRTELISGTKET---AATKPAEPEDIAAAIVKTLDKPKT
MLBr_00862|M.leprae_Br4923 GVGLTTICPGVIDTNIIQSTRIDTPTAKQERVRDRRGQLDMMFRLRRYGP
: . : * . *:: . . :: : .
MAB_4335|M.abscessus_ATCC_1997 PVFVPKMFGGMYRTMSVLPRSTGKFLTRKLGIQDFMLNAHGSDARKAYED
Mvan_2345|M.vanbaalenii_PYR-1 TVYVPARLRPVSWAYGLLPGAARTQVMAALGAGHRMLDGD-DTARRDYNR
MAV_2700|M.avium_104 RKTVPATLGALLKSVSLLPDTPRFAVAHAVRF-DRLVSGADPDARAAYHR
Mb1896c|M.bovis_AF2122/97 RVRVTKAAGSMIVAQRFMPRQVSEGLNRLLGGEHVFTDDVDMEKRRTYEA
Rv1865c|M.tuberculosis_H37Rv RVRVTKAAGSMIVAQRFMPRQVSEGLNRLLGGEHVFTDDVDMEKRRTYEA
MMAR_2743|M.marinum_M RVRVTKAAGAMIASQKYLPRWLSEGLNRLLGGEHVFTDDVDAAKREAYEA
MUL_3011|M.ulcerans_Agy99 RVRVTKAAGAMIASQKYLPRWLSEGLNRLLGGEHVFTDDVDAAKREAYEA
Mflv_0217|M.gilvum_PYR-GCK HVSVPPPLRFTAQAAQMLPPKGRRAMNKALGLDTVFLD-VDTDKRKGYEE
MSMEG_0771|M.smegmatis_MC2_155 HVSVPPPLRFTAQAAQMLPPKGRRWLNKKLGLDNVFLE-FDTAKRKAYED
TH_3427|M.thermoresistible__bu HVSVPGPLRFIAQAAQTLGPRGRRWLNKRLGLDTVFLE-FDQQARQGYEQ
MLBr_00862|M.leprae_Br4923 DKVADTILSAIKKNKPIRP--VAPEAYALYGISRVLPQVLRSTARIRVI-
. : . *
MAB_4335|M.abscessus_ATCC_1997 RASHSGPSTE-------------
Mvan_2345|M.vanbaalenii_PYR-1 RIDTR------------------
MAV_2700|M.avium_104 RLAEES-----------------
Mb1896c|M.bovis_AF2122/97 RARGEE-----------------
Rv1865c|M.tuberculosis_H37Rv RARGEE-----------------
MMAR_2743|M.marinum_M RARGETGES--------------
MUL_3011|M.ulcerans_Agy99 RARGETGES--------------
Mflv_0217|M.gilvum_PYR-GCK RARAARGVVEGSQG---------
MSMEG_0771|M.smegmatis_MC2_155 RAQAALGVVESEKK---------
TH_3427|M.thermoresistible__bu RAQQALGVLEGGNKQQDRSGQKS
MLBr_00862|M.leprae_Br4923 -----------------------