For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VPGRTSIGVKIRDKVQDKVIAITGGARGIGLATAAALHNLGAKVAIGDIDEAMAKESGADLDLDMYGKLD VTDPDSFSGFLDAVERQLGPIDVLVNNAGIMPVGRIVDEPDPVTRRILDINVYGVILGSKLAAQRMVPRG RGHVINVASLAGEIYAVGVATYCASKHAVVAFTDSARLEYRSAGVKFSMVLPSFVNTELIAGTGGIKGFK NAEPADIADAIVGLIVHPKPRVRVTKAAGSMIVAQRFMPRQVSEGLNRLLGGEHVFTDDVDMEKRRTYEA RARGEE
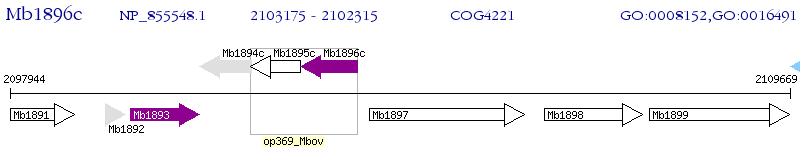
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1896c | - | - | 100% (286) | short chain dehydrogenase |
| M. bovis AF2122 / 97 | Mb2237c | ephD | 2e-33 | 32.95% (258) | short chain dehydrogenase |
| M. bovis AF2122 / 97 | Mb2025 | fabG3 | 1e-27 | 38.25% (183) | 20-beta-hydroxysteroid dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0217 | - | 2e-53 | 46.01% (276) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv1865c | - | 1e-160 | 100.00% (286) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00862 | ephD | 8e-31 | 36.10% (205) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4335 | - | 3e-56 | 45.86% (266) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_2743 | - | 1e-129 | 81.14% (281) | short-chain type dehydrogenase |
| M. avium 104 | MAV_2854 | - | 1e-125 | 78.65% (281) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0771 | - | 1e-49 | 42.60% (277) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3427 | - | 2e-46 | 41.67% (276) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3011 | - | 1e-128 | 80.07% (281) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_2345 | - | 2e-52 | 45.45% (264) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1896c|M.bovis_AF2122/97 --------------------------------------------------
Rv1865c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2743|M.marinum_M --------------------------------------------------
MUL_3011|M.ulcerans_Agy99 --------------------------------------------------
MAV_2854|M.avium_104 --------------------------------------------------
Mflv_0217|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0771|M.smegmatis_MC2_155 --------------------------------------------------
TH_3427|M.thermoresistible__bu --------------------------------------------------
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_2345|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIVLVHGFPDSHVLWD
Mb1896c|M.bovis_AF2122/97 --------------------------------------------------
Rv1865c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2743|M.marinum_M --------------------------------------------------
MUL_3011|M.ulcerans_Agy99 --------------------------------------------------
MAV_2854|M.avium_104 --------------------------------------------------
Mflv_0217|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0771|M.smegmatis_MC2_155 --------------------------------------------------
TH_3427|M.thermoresistible__bu --------------------------------------------------
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_2345|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVVPLLAKRFRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDELSP
Mb1896c|M.bovis_AF2122/97 --------------------------------------------------
Rv1865c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2743|M.marinum_M --------------------------------------------------
MUL_3011|M.ulcerans_Agy99 --------------------------------------------------
MAV_2854|M.avium_104 --------------------------------------------------
Mflv_0217|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0771|M.smegmatis_MC2_155 --------------------------------------------------
TH_3427|M.thermoresistible__bu --------------------------------------------------
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_2345|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSG
Mb1896c|M.bovis_AF2122/97 --------------------------------------------------
Rv1865c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2743|M.marinum_M --------------------------------------------------
MUL_3011|M.ulcerans_Agy99 --------------------------------------------------
MAV_2854|M.avium_104 --------------------------------------------------
Mflv_0217|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0771|M.smegmatis_MC2_155 --------------------------------------------------
TH_3427|M.thermoresistible__bu --------------------------------------------------
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_2345|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMV
Mb1896c|M.bovis_AF2122/97 --------------------------------------------------
Rv1865c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2743|M.marinum_M --------------------------------------------------
MUL_3011|M.ulcerans_Agy99 --------------------------------------------------
MAV_2854|M.avium_104 --------------------------------------------------
Mflv_0217|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0771|M.smegmatis_MC2_155 --------------------------------------------------
TH_3427|M.thermoresistible__bu --------------------------------------------------
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_2345|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQ
Mb1896c|M.bovis_AF2122/97 --------------------------------------------------
Rv1865c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2743|M.marinum_M --------------------------------------------------
MUL_3011|M.ulcerans_Agy99 --------------------------------------------------
MAV_2854|M.avium_104 --------------------------------------------------
Mflv_0217|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0771|M.smegmatis_MC2_155 --------------------------------------------------
TH_3427|M.thermoresistible__bu --------------------------------------------------
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_2345|M.vanbaalenii_PYR-1 -----------------------------------------------MVP
MLBr_00862|M.leprae_Br4923 LIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAAVHD
Mb1896c|M.bovis_AF2122/97 -----------MPGRTSIGVKIRDKVQDKVIAITGGARGIGLATAAALHN
Rv1865c|M.tuberculosis_H37Rv -----------MPGRTSIGVKIRDKVQDKVIAITGGARGIGLATAAALHN
MMAR_2743|M.marinum_M -----------MADSTTIGVRVR----DKVIVITGGARGIGLATATALHK
MUL_3011|M.ulcerans_Agy99 -----------MADSTTIGVRVR----NKVIVITGGARGIGLATATALHK
MAV_2854|M.avium_104 -----------MADTASIGLKVR----DKVVVITGGARGIGLATATALHK
Mflv_0217|M.gilvum_PYR-GCK -----------MDN-------IR----GKTIAITGAARGIGYATAEALLA
MSMEG_0771|M.smegmatis_MC2_155 -----------MDN-------IR----GKTIAITGAARGIGYATAKALLA
TH_3427|M.thermoresistible__bu -----------MDN-------IR----GKTIAITGAARGIGYATATALLA
MAB_4335|M.abscessus_ATCC_1997 -------------------MGTAVQLAGKVVAVTGGARGIGRAIATAFAA
Mvan_2345|M.vanbaalenii_PYR-1 SRPARGPRTSAQLGMKGGTVTADRNLTGRVVAITGAARGIGAATARAFAA
MLBr_00862|M.leprae_Br4923 LADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAGSGIGRETALALAR
. : :**.. *** * *:
Mb1896c|M.bovis_AF2122/97 LGAKVAIGDIDEAMAKESGADLD--LDMYG--KLDVTDPDSFSGFLDAVE
Rv1865c|M.tuberculosis_H37Rv LGAKVAIGDIDEAMAKESGADLD--LDMYG--KLDVTDPDSFSGFLDAVE
MMAR_2743|M.marinum_M LGAKVAIGDVDEPAVKEAGADLG--LEVYG--KLDVTDPNSFSDFLDQVE
MUL_3011|M.ulcerans_Agy99 LGAKVAIGDVGEPAVKEAGADLG--LEVYG--KLDVTDPNSFSDFLDQVE
MAV_2854|M.avium_104 LGAKVAIGDIDEVRVKESGAALD--LDVYG--KLDVTDPHSFSDFLDEVE
Mflv_0217|M.gilvum_PYR-GCK HGARVVIGDRDVALQESSVAKLTN-LGPVSGYPLDVTDQESFATFLDKAR
MSMEG_0771|M.smegmatis_MC2_155 HGARVVIGDRDVALQESAVVELSK-LGQVSGYPLDVTDRESFATFLDKAR
TH_3427|M.thermoresistible__bu RGARVVIGDRDVALQESAVAQLGR-LGPVSGYPLDVTDRESFATFLDKAR
MAB_4335|M.abscessus_ATCC_1997 EGAKVAIGDIDKKLCENTAAEIGN-SAIGLP--LDVTDHGSFEAFFDTIA
Mvan_2345|M.vanbaalenii_PYR-1 AGASVAIGDLDADLAVRTAAEIG---CLGSG--VDVTDHDSVEKFLDEVE
MLBr_00862|M.leprae_Br4923 RGAEVVVSDIDEAAAKNTAAQIVASGGVAYAYALDVSDAAAVEAFAEQVS
** *.:.* . : . : :**:* :. * :
Mb1896c|M.bovis_AF2122/97 RQLG-PIDVLVNNAGIMPVGRIVDEPDPVTRRILDINVYGVILGSKLAAQ
Rv1865c|M.tuberculosis_H37Rv RQLG-PIDVLVNNAGIMPVGRIVDEPDPVTRRILDINVYGVILGSKLAAQ
MMAR_2743|M.marinum_M RQLG-PLDVLVNNAGIMPVGRIVDEPDSVTRRILDINVYGVMVGSKLAAQ
MUL_3011|M.ulcerans_Agy99 RQLG-PLDVLVNNAGIMPVGRIVDEPDSVTRRILDINVYGVMVGSKLAAQ
MAV_2854|M.avium_104 RQLG-PIDVLVNNAGIMPLGRVVDESDAVTRRILDINVYGVILGSKLALA
Mflv_0217|M.gilvum_PYR-GCK TDGGGHIDVLINNAGVMPVGPFLDETEQSIRSSLEVNVYGVLTGCRLALP
MSMEG_0771|M.smegmatis_MC2_155 TDGGGHIDVLINNAGVMPVGPFLEQSEQSIRSNIEVNVYGVLTGCQLALP
TH_3427|M.thermoresistible__bu TDGGGHIDVLINNAGVMPIGPFLEQSEQAIRSSIEVNLYGVLTGCQLALP
MAB_4335|M.abscessus_ATCC_1997 ATVG-PVDVIVNNAGIMPITPFDDESLESIQRQLDINVRGVMWGSQLAIK
Mvan_2345|M.vanbaalenii_PYR-1 QRLG-PLDVMVNNAGIMPVTQMLDESPESVRRQLAVNVAGVVFGTQHAAS
MLBr_00862|M.leprae_Br4923 AKHG-VPDIVVNNAGIGQAGRFLDTPPEEFDRVLAVNLGGVVNCCRAFGQ
* *:::****: . : . : :*: **: :
Mb1896c|M.bovis_AF2122/97 RMVPRGRG-HVINVASLAGEIYAVGVATYCASKHAVVAFTDSARLEYRSA
Rv1865c|M.tuberculosis_H37Rv RMVPRGRG-HVINVASLAGEIYAVGVATYCASKHAVVAFTDSARLEYRSA
MMAR_2743|M.marinum_M RMVPRGRG-HVINVASLAGEIYVVGLATYCASKHAVIAFTDAARIEYRST
MUL_3011|M.ulcerans_Agy99 RMVPRGRG-HVINVASLAGEIYVVGLATYCASKHAVIAFTDAARIEYRTT
MAV_2854|M.avium_104 RMIPRGRG-HVINVASLAGETYLAGAATYCASKHAVVGFTDAARIEYRRS
Mflv_0217|M.gilvum_PYR-GCK DMVKRRSG-HIINIASLSGVIPLPGQVVYVGAKYAVVGLTTALADEMAPH
MSMEG_0771|M.smegmatis_MC2_155 DMVKRRRG-HIINIASLSGVIPLPGQVVYVGAKYAVVGLSTALADEMAPY
TH_3427|M.thermoresistible__bu DMVRRRSG-HIINIASMSGLIPVPGQVPYNAAKFGVVGLSVALADEMAPY
MAB_4335|M.abscessus_ATCC_1997 RMKPRGGG-VIVNIASAAGKAGFPGLATYCATKHAVVGLSESLSLEYEPA
Mvan_2345|M.vanbaalenii_PYR-1 RMVARRSG-HIVNVASAAGRIAFGGVATYTATKFAVVGFTEAAALELRDS
MLBr_00862|M.leprae_Br4923 RLVERGTGGHIVNVSSMAAYAPLQSLSAYCTSKAATYMFSDCLRAELDAV
: * * ::*::* :. . * :* .. :: . *
Mb1896c|M.bovis_AF2122/97 GVKFSMVLPSFVNTELIAGTGGIKGFKNAEPADIADAIVGLIVHPKPRVR
Rv1865c|M.tuberculosis_H37Rv GVKFSMVLPSFVNTELIAGTGGIKGFKNAEPADIADAIVGLIVHPKPRVR
MMAR_2743|M.marinum_M GVKFSMVLPTFVNTELASGTPGMKGFKNAEPSDIADAIVALVANPKPRVR
MUL_3011|M.ulcerans_Agy99 GVKFSMVLPTFVNTELASGTPGMKGFKNAEPSDIADAIVALVANPKPRVR
MAV_2854|M.avium_104 GVKFSVVKPTFVNTELIAGTSGAKGVRNAEPSDIADAIVKLVAHPRPRVR
Mflv_0217|M.gilvum_PYR-GCK GVNVSVIMPPFTNTELIAGTKSGGAIKPVEPEDIAAAIVKTLNKPKTHVS
MSMEG_0771|M.smegmatis_MC2_155 GVDVSVIMPPFTNTDLISGTKSGGAIKPVEPEEIAAAIVKTLNKPKTHVS
TH_3427|M.thermoresistible__bu GVEVSVVMPPFTRTELISGTKETAATKPAEPEDIAAAIVKTLDKPKTHVS
MAB_4335|M.abscessus_ATCC_1997 GISVVCVMPGMVNTELISGLDTHWLLKTVEPEDIANAVVKAVRRGVFPVF
Mvan_2345|M.vanbaalenii_PYR-1 GVRFTCVLPGVVNTELTSGLRDHWLLRSCEPEEVAAGVVAGVRRHRRTVY
MLBr_00862|M.leprae_Br4923 GVGLTTICPGVIDTNIIQSTR-------IDTPTAKQERVRDRRGQLDMMF
*: . : * . *:: . :. * :
Mb1896c|M.bovis_AF2122/97 VTKAAGSMIVAQRFMPRQVSEGLNRLLGGEHVFTDDVDMEKRRTYEARAR
Rv1865c|M.tuberculosis_H37Rv VTKAAGSMIVAQRFMPRQVSEGLNRLLGGEHVFTDDVDMEKRRTYEARAR
MMAR_2743|M.marinum_M VTKAAGAMIASQKYLPRWLSEGLNRLLGGEHVFTDDVDAAKREAYEARAR
MUL_3011|M.ulcerans_Agy99 VTKAAGAMIASQKYLPRWLSEGLNRLLGGEHVFTDDVDAAKREAYEARAR
MAV_2854|M.avium_104 VTRTAGAIIASQKFMPRALSEGLNRLLGGEHVFTDAVDVEKRQAYEARAR
Mflv_0217|M.gilvum_PYR-GCK VPPPLRFTAQAAQMLPPKGRRAMNKALGLDTVFLD-VDTDKRKGYEERAR
MSMEG_0771|M.smegmatis_MC2_155 VPPPLRFTAQAAQMLPPKGRRWLNKKLGLDNVFLE-FDTAKRKAYEDRAQ
TH_3427|M.thermoresistible__bu VPGPLRFIAQAAQTLGPRGRRWLNKRLGLDTVFLE-FDQQARQGYEQRAQ
MAB_4335|M.abscessus_ATCC_1997 VPKMFGGMYRTMSVLPRSTGKFLTRKLGIQDFMLNAHGSDARKAYEDRAS
Mvan_2345|M.vanbaalenii_PYR-1 VPARLRPVSWAYGLLPGAARTQVMAALGAGHRMLDGD-DTARRDYNRRID
MLBr_00862|M.leprae_Br4923 RLRRYGPDKVADTILSAIKKNKPIRPVAPEAYALYGISRVLPQVLRSTAR
: : :. . .
Mb1896c|M.bovis_AF2122/97 GEE-----------------
Rv1865c|M.tuberculosis_H37Rv GEE-----------------
MMAR_2743|M.marinum_M GETGES--------------
MUL_3011|M.ulcerans_Agy99 GETGES--------------
MAV_2854|M.avium_104 GEQ-----------------
Mflv_0217|M.gilvum_PYR-GCK AARGVVEGSQG---------
MSMEG_0771|M.smegmatis_MC2_155 AALGVVESEKK---------
TH_3427|M.thermoresistible__bu QALGVLEGGNKQQDRSGQKS
MAB_4335|M.abscessus_ATCC_1997 HSGPSTE-------------
Mvan_2345|M.vanbaalenii_PYR-1 TR------------------
MLBr_00862|M.leprae_Br4923 IRVI----------------