For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPKTFFITGVSSGLGRALAHGALEAGHTVIGTVRKTADLAPFEALDAKAHGRILDVTDHAAVIDTVAEAE ASIGPIDVLIANAGYGLEGVFEETPLSEVRAQFETNFFGAAATVQSVLPYMRKRRGGHIMAVTSMGGLMT VPGLSVYCASKYALEGLLESIGKEVAGFGIHVTAIEPGSFRTDWAGRSMVRAERAIDDYDAVFEPIRSAR LRANGNQLGDPAKAATAILTAVEAAQPPRHLLLGSDALRLIRAGRDAVDADIAAWESLSRSTDFPDGHQI AAS
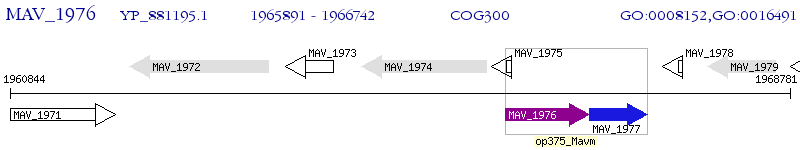
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1976 | - | - | 100% (283) | short chain dehydrogenase |
| M. avium 104 | MAV_4982 | - | 3e-53 | 42.96% (277) | short-chain dehydrogenase/reductase SDR |
| M. avium 104 | MAV_3827 | - | 6e-52 | 42.86% (273) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3112 | - | 1e-20 | 28.73% (268) | short-chain type dehydrogenase/reductase |
| M. gilvum PYR-GCK | Mflv_0295 | - | 2e-48 | 41.07% (280) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv3085 | - | 1e-20 | 28.73% (268) | short-chain type dehydrogenase/reductase |
| M. leprae Br4923 | MLBr_01094 | - | 1e-18 | 26.58% (222) | short chain alcohol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1305 | - | 2e-25 | 37.43% (179) | short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_1614 | - | 5e-56 | 43.91% (271) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3426 | - | 1e-112 | 69.86% (282) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3670 | - | 4e-47 | 42.35% (255) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3423 | - | 8e-56 | 43.54% (271) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0982 | - | 2e-50 | 42.12% (273) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_1976|M.avium_104 ------------MPKTFFITGVSSGLGRALAHGALEAGHTVIGTVRKTAD
MSMEG_3426|M.smegmatis_MC2_155 -------------MKTFFITGVSSGLGQAFANGALAAGHRVVGTVRKDSD
MMAR_1614|M.marinum_M -------------MSTWLITGCSTGLGRALAEAVIDAGHHTVATARSVGG
MUL_3423|M.ulcerans_Agy99 -------------MSTWLITGCSTGLGRALAEAVIDAGHHTVATARSVGG
TH_3670|M.thermoresistible__bu MGSNVWG------SKVWFITGASRGFGREWAIAALDRGDRVAATARDVST
Mvan_0982|M.vanbaalenii_PYR-1 MPGHPTGIDPGMTEKVWFITGASRGFGREWAIAALDRGDKVAATARDLST
Mflv_0295|M.gilvum_PYR-GCK ------------MSKVLLISGASRGLGRAITESALAAGHLVVAGVRATSS
Mb3112|M.bovis_AF2122/97 --------MSSFEGKVAVITGAGSGIGRALALNLSEKRAKLALSDVDTDG
Rv3085|M.tuberculosis_H37Rv --------MSSFEGKVAVITGAGSGIGRALALNLSEKRAKLALSDVDTDG
MLBr_01094|M.leprae_Br4923 --------MEGFAGKVAVVTGAGSGIGQALAIELARSGAKLAISDVDGEG
MAB_1305|M.abscessus_ATCC_1997 ----------MSEHWTVLVTGASSGIGASAARIFAERGHRVFGTSRNPDT
. .::* . *:* :
MAV_1976|M.avium_104 LAP---FEALD-AKAHGRILDVTDHAAVIDTVAEAEASIGPIDVLIANAG
MSMEG_3426|M.smegmatis_MC2_155 AAA---FESTAPDRAHARQLDVTDDAAVTAVVAEVEKNIGPIDVVIANAG
MMAR_1614|M.marinum_M VAD---LVQRSPERVLPLALDITEPDQITAAVQAAQQRFGGIDVLVNNAG
MUL_3423|M.ulcerans_Agy99 VAD---LAQRSPERVLPLALDITEPDQITAAMQAAQQRFGGIDVLVNNAG
TH_3670|M.thermoresistible__bu LDD---LRDRFGDAVLAMTLDVTDRAADFAAVQRAHDHFGRLDIVVNNAG
Mvan_0982|M.vanbaalenii_PYR-1 LDD---LTAKYGDALLPLTLDVTDRDADFAAVTQAHDHFGRLDIVVNNAG
Mflv_0295|M.gilvum_PYR-GCK LSD---LATREPDRLAVVELDVTDDRHARAAVATAVERFGRLDVLVNNAG
Mb3112|M.bovis_AF2122/97 LAKTVRLAQALGAQVKSDRLDVAEREAVLAHADAVVAHFGTVHQVYNNAG
Rv3085|M.tuberculosis_H37Rv LAKTVRLAQALGAQVKSDRLDVAEREAVLAHADAVVAHFGTVHQVYNNAG
MLBr_01094|M.leprae_Br4923 LAQTEGQLKAIGASARTDRLDVTEREAFLTYADVVHENFGKVNQIYNNAG
MAB_1305|M.abscessus_ATCC_1997 ITD-------RVPGVQYLRLDQTDK----ASIAECVAQAGEVDILVNNAG
** :: * :. : ***
MAV_1976|M.avium_104 YGLEGVFEETPLSEVRAQFETNFFGAAATVQSVLPYMRKRRGGHIMAVTS
MSMEG_3426|M.smegmatis_MC2_155 YGVEGIFEETPIAEMRAQFATNVFGVAATLQAALPYLRERRKGHLMAVTS
MMAR_1614|M.marinum_M YGYRAAVEEGDDAEVRDLFETHFFGTVALIKAVLPDMRARRSGAIVNISS
MUL_3423|M.ulcerans_Agy99 YGYRAAVEEGDDAEVRDLFETHFFGTVALIKAVLPDMRARRSGAIVNISS
TH_3670|M.thermoresistible__bu YGHFGFVEELTETEVRDQMETNFFGALWVTQAALPYLRRQRSGHIIQVSS
Mvan_0982|M.vanbaalenii_PYR-1 YGQFGFVEELSEQEARAQIDTNVFGALWITQAALPYLRAQRSGHLIQVSS
Mflv_0295|M.gilvum_PYR-GCK YANMSAIEDVDVDDFRAQIETNFFGVVNLTRAALPAMRRQRDGHIVQISS
Mb3112|M.bovis_AF2122/97 IAYNGNVDKSEFKDIERIIDVDFWGVVNGTKAFLPHVIASGDGHIVNISS
Rv3085|M.tuberculosis_H37Rv IAYNGNVDKSEFKDIERIIDVDFWGVVNGTKAFLPHVIASGDGHIVNISS
MLBr_01094|M.leprae_Br4923 IAFTGDVEVSHFKDIERVMDVDYWGVVNGTKAFLPYLISSGDGHVINISS
MAB_1305|M.abscessus_ATCC_1997 ESQIGALEDVSMDDFEKLYMTNVFGPVALTKAVLPGMRERRRGAVVMVGS
. . .: : . .. :* :: ** : * :: : *
MAV_1976|M.avium_104 MGGLMTVPGLSVYCASKYALEGLLESIGKEVAGFG--IHVTAIEPGSFRT
MSMEG_3426|M.smegmatis_MC2_155 MGGLMAVPGMSAYCASKFAVEGMLESIRKEISGFG--IHVTAIEPGSFRT
MMAR_1614|M.marinum_M IAVALTPVGSGYYAAAKAAMEGMSGALHGELAPLG--ISVTVVEPGAFRT
MUL_3423|M.ulcerans_Agy99 IAVALTPVGSGYYAAAKAAMEGMSGALHGELAPLG--ISVTVVEPGAFRT
TH_3670|M.thermoresistible__bu IGGVSAFADLGAYHASKWALEGLSQSLSLEVAPFG--IHVTLIEPGGYAT
Mvan_0982|M.vanbaalenii_PYR-1 IGGIVAFPNLGIYHASKWALEGFSQALAQEVAEFG--IHVTLIEPGGYST
Mflv_0295|M.gilvum_PYR-GCK VGGRLVRPGLAAYQSAKWAVTGFSGVLAVEVAPLG--IKVTVLEPGGMRT
Mb3112|M.bovis_AF2122/97 LFGLIAVPGQSAYNAAKFAVRGFTEALRQEMLVARHPVKVTCVHPGGIKT
Rv3085|M.tuberculosis_H37Rv LFGLIAVPGQSAYNAAKFAVRGFTEALRQEMLVARHPVKVTCVHPGGIKT
MLBr_01094|M.leprae_Br4923 VFGLFSVPGQAAYNSAKFAVRGFTEALREEMALAGRPVNVTTVYPGGIKT
MAB_1305|M.abscessus_ATCC_1997 MAGTLHVPFRSTYSSAKSALHTVADCLRYEVAPFG--ITVSTVEPGVIAT
: . * ::* *: . : *: : *: : ** *
MAV_1976|M.avium_104 DWAGRSMVRAERAIDDYDAVFEPIRSARLRANGNQLGDPAKAATAILTAV
MSMEG_3426|M.smegmatis_MC2_155 DWAGRSMTRADRSIADYDELFEPIRAARLKASGNQLGNPVKAAEAVLAIL
MMAR_1614|M.marinum_M DFAGRSLVQSTRMINDYAATAGQRRKENDTMHGNQPGDPAKAAAAIITAV
MUL_3423|M.ulcerans_Agy99 DFAGRSLVQSTRMINDYAATAGQRRKENDTMHGNQPGDPAKAAAAIITAV
TH_3670|M.thermoresistible__bu DWGGSSARRSA-TMPEYAESRAEADRVR-SQRVARPGDPKASAAALLKVV
Mvan_0982|M.vanbaalenii_PYR-1 DWAGASAKRAT-ALPAYAEAHAATERTR-QARTARPGDPQASARALLTVV
Mflv_0295|M.gilvum_PYR-GCK DWAGSSMRIAP-VREEYAATVGAAASLS-DPNVLGASDPAKVAGLLLDVI
Mb3112|M.bovis_AF2122/97 AVARNATVADGEDQQTFAEFFDR---------RLALHSPEMAAKTIVNGV
Rv3085|M.tuberculosis_H37Rv AVARNATVADGEDQQTFAEFFDR---------RLALHSPEMAAKTIVNGV
MLBr_01094|M.leprae_Br4923 AIARNATAAEGLDVSKIASRFDT---------WVAHTSPQHAARIILKAV
MAB_1305|M.abscessus_ATCC_1997 GIETRRNRIVPDGSPYRDAFRTVDAAMA--AREEHGTATDELGRLVVDVA
. . ::
MAV_1976|M.avium_104 EAAQPPRHLLLGSDALRLIRAGRDAVDADIAAWESLSRSTDFPDGHQIAA
MSMEG_3426|M.smegmatis_MC2_155 DEPEPPAHLVLGSDALRLVGAARAAVDEDIRRWEELSRSTDFADGAQLAS
MMAR_1614|M.marinum_M ESPAPPAFLLLGPDALMAYRHIAQRRDVEITDWEQLTAGTNLTP------
MUL_3423|M.ulcerans_Agy99 ESPAPPAFLLLGPDALMAYRHIAQRRDVEITDWEQLTAGTNLTP------
TH_3670|M.thermoresistible__bu DAEKPPLRVFFGELPLQIVNADYQSRLQTWEEWQPIAVLAQG--------
Mvan_0982|M.vanbaalenii_PYR-1 DAPEPPLRVFFGELPLELARADYESRLANWEKWQPVAIEAQG--------
Mflv_0295|M.gilvum_PYR-GCK GMPDPPRRLLVGPDAYRYATAAGRDLLAVDERWRTLSMSTAADDVSPEQL
Mb3112|M.bovis_AF2122/97 AKGQ--ARVVVGLEAKAVDVLARIMG-SSYQRLVAAGVAKFFPWAK----
Rv3085|M.tuberculosis_H37Rv AKGQ--ARVVVGLEAKAVDVLARIMG-SSYQRLVAAGVAKFFPWAK----
MLBr_01094|M.leprae_Br4923 RKKK--ARVLVGPDAKVANVVVRFSGGAGYQRLFAQVASRLILNQR----
MAB_1305|M.abscessus_ATCC_1997 LSPSPKPLYAKGSNAPIIMTAQRLLPARLFLKFLARMHGLKVR-------
* .
MAV_1976|M.avium_104 S-------
MSMEG_3426|M.smegmatis_MC2_155 T-------
MMAR_1614|M.marinum_M --------
MUL_3423|M.ulcerans_Agy99 --------
TH_3670|M.thermoresistible__bu --------
Mvan_0982|M.vanbaalenii_PYR-1 --------
Mflv_0295|M.gilvum_PYR-GCK DPLGSASR
Mb3112|M.bovis_AF2122/97 --------
Rv3085|M.tuberculosis_H37Rv --------
MLBr_01094|M.leprae_Br4923 --------
MAB_1305|M.abscessus_ATCC_1997 --------