For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PSRTYGGATSGERRARRRAALLDAALDLVAGSGVKGLTVRGVCAEARLNDRYFYESFRSTDELVLAMLDE QMLAGSATIIEAIAKSSTSKDPVVRTRAAVGSGLRFLTADPRRGRLLVESQATEGLAARRRDLVKTLAQV MADQGRVLLPADDALDPDLDLATFTLVSGGLDLVTTWFRGELDITRDRLEDFLVALVTRANIRADQHEDS GEAAKPGN*
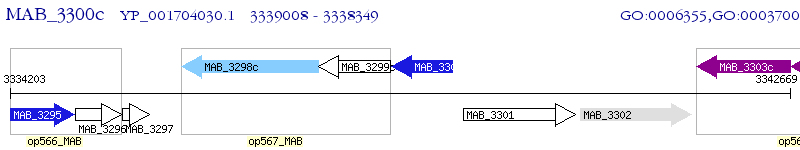
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3300c | - | - | 100% (219) | TetR family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_1638 | - | 4e-24 | 35.86% (198) | TetR family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_2070c | - | 3e-21 | 32.98% (188) | putative transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0281c | - | 7e-19 | 32.99% (197) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_1526 | - | 2e-23 | 34.34% (198) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0275c | - | 3e-18 | 32.49% (197) | TetR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_3142 | - | 1e-20 | 32.43% (185) | TetR family transcriptional regulator |
| M. avium 104 | MAV_3986 | - | 4e-25 | 38.00% (200) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_1355 | - | 4e-24 | 36.87% (198) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_4000 | - | 5e-24 | 37.25% (204) | PUTATIVE transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_1200 | - | 3e-19 | 30.00% (200) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_2901 | - | 1e-33 | 41.62% (197) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_3300c|M.abscessus_ATCC_199 -----MPSRTYGGATSGERRARRRAALLDAALDLVAGS--GVKGLTVRGV
Mvan_2901|M.vanbaalenii_PYR-1 -------MALHRGLTADQRREQRRQQLIEAALDVIAEG--GVGGLRVRAI
Mflv_1526|M.gilvum_PYR-GCK --MTDAPRRTYGGQSADARRLARRARLLDAAIEVIADN--AWRTLTVDRL
Mb0281c|M.bovis_AF2122/97 ---MTRSDRPYRGVEAAERLATRRRQLLSAGLDLLGSDQHDIAELTIRTI
Rv0275c|M.tuberculosis_H37Rv ---MTRSDRPYRGVEAAERLATRRRQSLSAGLDLLGSDQHDIAELTIRTI
MUL_1200|M.ulcerans_Agy99 -------MRPYRGVEATERLATRRSQLLESGLELLGSSDRDIAELTVRGI
MSMEG_1355|M.smegmatis_MC2_155 ----MTTPTRWAGVPLTDRRAERRALLVDAAFRLFGDG--GETALSVRSV
TH_4000|M.thermoresistible__bu ----MTSPTRWAGVPLTDRRAERRALLIAAAYRLFGEG--GEAALSVRSV
MAV_3986|M.avium_104 ----MSSPSRWAGVPLKDRRAERRALLVEAAYRLFGSE--GEAAVSVRSV
MMAR_3142|M.marinum_M MFDVRSRNTRWGGRTGAERRAERRQQLIEAATEIWSES--GWAAVTMRGV
* * ** : :. : . : : :
MAB_3300c|M.abscessus_ATCC_199 CAEARLNDRYFYESFRSTDELVLAMLDEQMLAGSATIIEAIAK------S
Mvan_2901|M.vanbaalenii_PYR-1 SERARLNDRYFYESFRDCQELMLAAYDDQFARALTGVTAVIAE------S
Mflv_1526|M.gilvum_PYR-GCK CADAGLNKRYFYESFATLDDVATAVVDDISEDVRTATMDVAAQ------R
Mb0281c|M.bovis_AF2122/97 CRRAGLSVRYFYESFTDKDEFVGRVFDWVVAELVATTQAAVTA------V
Rv0275c|M.tuberculosis_H37Rv CRRAGLSVRYFYESFTDKDEFVGRVFDWVVAELVATTQAAVTA------V
MUL_1200|M.ulcerans_Agy99 CRQAGLSARYFYESFPDKDEFIGGVFDWVVGDLATTIQAAVAT------V
MSMEG_1355|M.smegmatis_MC2_155 CRECGLNTRYFYESFADTDDLIGAVYDEVSALLAADVEAAM--------E
TH_4000|M.thermoresistible__bu CRESELNSRYFYESFADVDELLGAVYDEVSAQLAAEVETAMAAAVEGAGE
MAV_3986|M.avium_104 CRECGLNTRYFYESFRDTDDLLGAVYDRVSEQLEEVIAAAI--------E
MMAR_3142|M.marinum_M CARTGLNDRYFYEDFKTREDLLVAAWDGVRNDMLGEVSALFDER------
. *. *****.* ::. *
MAB_3300c|M.abscessus_ATCC_199 STSKDPVVRTRAAVGSGLRFLTADPRRGRLLVESQA-TEGLAARRRDLVK
Mvan_2901|M.vanbaalenii_PYR-1 PP--ELKLRVREILEFAIGFLDEDPRRSRLLIELQT-SEALAERRHEVLG
Mflv_1526|M.gilvum_PYR-GCK AD-EPLDQQARAVVDALVRTLVEDPRRGRILLGGVAGSVALHEHRGAVMR
Mb0281c|M.bovis_AF2122/97 PA----REQTRAGMANIVRTITADARVGRLLFSTQLANAVITRKRAESSA
Rv0275c|M.tuberculosis_H37Rv PA----REQTRAGMANIVRTITADARVGRLLFSTQLANAVITRKRAESSA
MUL_1200|M.ulcerans_Agy99 PP----PEQARAGIAEIVRTIVGDARIGRLLFSTHLANPVVVRKRTESSA
MSMEG_1355|M.smegmatis_MC2_155 GAGNSLRARTRAGIAAVLGFSSADPRRGRVLFTDARANPVLAQRRAVTQD
TH_4000|M.thermoresistible__bu GAGDSLRARTRAGIVAVLGFSSADPRRGRILFTDARANPVLAARRAATQD
MAV_3986|M.avium_104 QAGDSLRARTRAGIAAVLGFSSADPRRGRVLFTDARTNPVLAARRRATQD
MMAR_3142|M.marinum_M -VDRPPIETITAAIAIVVDRIARDPGRAHILLAQHVGSSPLQDRRAVALQ
: : *. .::*. . : :*
MAB_3300c|M.abscessus_ATCC_199 TLAQVMADQGRVLLPADDALDPDLDLATFTLVSGGLDLVTTWFRGELDIT
Mvan_2901|M.vanbaalenii_PYR-1 VLTQIMVG----LVGDGSGADDTATLTAVTVSSGLLELAAQWYREQIDIS
Mflv_1526|M.gilvum_PYR-GCK GLTAVLVSHARSVHGVELEKDPLAEVAPAFIVGGTADAILAYLDGRARMS
Mb0281c|M.bovis_AF2122/97 LFAMLSGQHAVDTLHAP--ANDHVKAVAHFAVGGVGQTISAWLAGDVRLD
Rv0275c|M.tuberculosis_H37Rv LFAMLSGQHAVDTLHAP--ANDHVKAVAHFAVGGVGQTISAWLAGDVRLD
MUL_1200|M.ulcerans_Agy99 LFAMLSGQHAGDALRIP--ANDRIKVGAHFIVGGVAQTISAWIEGDVELE
MSMEG_1355|M.smegmatis_MC2_155 LLREAVLSEGGRLNPGS--DPVAAQVGAAMYTGAMAELAQQWLAGNLGDD
TH_4000|M.thermoresistible__bu QLREAVLTEGWRLHPKS--DPVAAQVGAAMYTGAMAELAQQWLAGNLGDD
MAV_3986|M.avium_104 MLRQGVLSEGWRLNPRS--DPVAAEVAAAMYTGAMAELAQQWLAGHLGGD
MMAR_3142|M.marinum_M EATQLVVEASRPHLRED-ADETALRMDTLVAVGGFVEVITAWHSGLLAVT
. .. : :
MAB_3300c|M.abscessus_ATCC_199 RDRLEDFLVALVTR-ANIR---ADQHED--SGEAAKPGN-----------
Mvan_2901|M.vanbaalenii_PYR-1 RAQLIEFATALVVTTADITGVLERQLAPPISGDSTDDGRS----------
Mflv_1526|M.gilvum_PYR-GCK LDSLVTALTTLWLITGNGAAEVARHRLDG---------------------
Mb0281c|M.bovis_AF2122/97 PDQLVDQLAALLDELTDPNLSRPRVAATAAKSGANDPQPPEVAGQPPSSA
Rv0275c|M.tuberculosis_H37Rv PDQLVDQLAALLDELTDPNLSRPRVAATAAKSGANDPQPPEVAGQPPSSA
MUL_1200|M.ulcerans_Agy99 PGQLVDQLASLLDELSDPRLYGLR----GTRAGATAASPAQ---------
MSMEG_1355|M.smegmatis_MC2_155 LDAVVDHALRLVLGR-----------------------------------
TH_4000|M.thermoresistible__bu LDRVVDYALRLVLGR-----------------------------------
MAV_3986|M.avium_104 LDAVVDYASKLVLR------------------------------------
MMAR_3142|M.marinum_M EKEVVAHTSRLAETLAQRYVVSG---------------------------
: *
MAB_3300c|M.abscessus_ATCC_199 ------
Mvan_2901|M.vanbaalenii_PYR-1 ------
Mflv_1526|M.gilvum_PYR-GCK ------
Mb0281c|M.bovis_AF2122/97 RPARRS
Rv0275c|M.tuberculosis_H37Rv RPARRS
MUL_1200|M.ulcerans_Agy99 ------
MSMEG_1355|M.smegmatis_MC2_155 ------
TH_4000|M.thermoresistible__bu ------
MAV_3986|M.avium_104 ------
MMAR_3142|M.marinum_M ------