For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PDHGRWNHNIHYQREVLRRVPAHATDVLDVGCGIGMLTRELAPLAQRVVGIDPHEPSLRIARAETSASNV EYVRGSFLEHPFPAESFDLIAAVAALHHMDIEQGLRRMSDLLKPGGRIVIVGFAANRSVTDLWYDGLGFW LHRYYRLRYGWWEHPSPVSCEGLDSHDEVRRIAQRVLPGARLQRLVLWRHLLTWEKS*
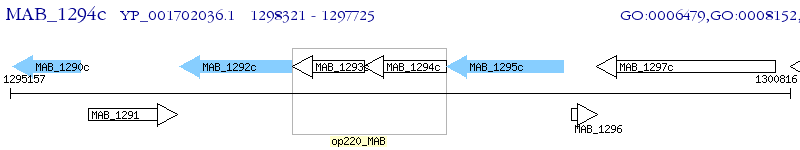
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1294c | - | - | 100% (198) | hypothetical protein MAB_1294c |
| M. abscessus ATCC 19977 | MAB_4018 | - | 9e-13 | 40.82% (98) | similarity with UbiE/COQ5 methyltransferase |
| M. abscessus ATCC 19977 | MAB_3689 | - | 3e-10 | 33.33% (102) | putative methyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0092 | - | 6e-24 | 33.51% (191) | methyltransferase/methylase |
| M. gilvum PYR-GCK | Mflv_4035 | - | 2e-12 | 40.00% (100) | methyltransferase type 11 |
| M. tuberculosis H37Rv | Rv0089 | - | 6e-24 | 33.51% (191) | methyltransferase/methylase |
| M. leprae Br4923 | MLBr_01723 | - | 6e-07 | 31.68% (101) | hypothetical protein MLBr_01723 |
| M. marinum M | MMAR_0244 | - | 3e-23 | 31.82% (198) | methyltransferase/methylase |
| M. avium 104 | MAV_4317 | - | 2e-09 | 32.35% (102) | putative methyltransferase |
| M. smegmatis MC2 155 | MSMEG_4708 | - | 1e-29 | 36.27% (204) | putative methyltransferase |
| M. thermoresistible (build 8) | TH_1968 | - | 6e-11 | 32.35% (102) | POSSIBLE METHYLTRANSFERASE (METHYLASE) |
| M. ulcerans Agy99 | MUL_4869 | - | 1e-22 | 31.31% (198) | methyltransferase/methylase |
| M. vanbaalenii PYR-1 | Mvan_4082 | - | 6e-28 | 35.68% (199) | methyltransferase type 12 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0092|M.bovis_AF2122/97 -----------------------------------------MDQPWNANI
Rv0089|M.tuberculosis_H37Rv -----------------------------------------MDQPWNANI
MMAR_0244|M.marinum_M ---------------------------------------MPAGQPWNINI
MUL_4869|M.ulcerans_Agy99 ---------------------------------------MPAGQPWNINI
MSMEG_4708|M.smegmatis_MC2_155 ---------------------------------------MPSPAYWNHNT
Mvan_4082|M.vanbaalenii_PYR-1 -----------------------------------------MQDYWNHNT
MAB_1294c|M.abscessus_ATCC_199 ---------------------------------------MPDHGRWNHNI
MAV_4317|M.avium_104 -------------------------MTRSRHDRSLSFGSAAAAYERGRPS
TH_1968|M.thermoresistible__bu ----------------------------------LSFGAEAAAYERGRPS
MLBr_01723|M.leprae_Br4923 MTRSTEISANAVPNPHATAEQVAAARKDSKLAQVLYHDWEAENYDEKWSI
Mflv_4035|M.gilvum_PYR-GCK -----------------MDGSERPVNHHGDHPGFSGLTGQLCAVAFLLTG
Mb0092|M.bovis_AF2122/97 HYDALLDAMVP--------------LGTQCVLDVGCGDGLLAARLARR--
Rv0089|M.tuberculosis_H37Rv HYDALLDAMVP--------------LGTQCVLDVGCGDGLLAARLARR--
MMAR_0244|M.marinum_M HYDALLAATVA--------------PGTQTALDVGCGDGFLSARLADR--
MUL_4869|M.ulcerans_Agy99 HYDALLAATVA--------------PGTQTALDVGCGDGFLSARLVDR--
MSMEG_4708|M.smegmatis_MC2_155 AYHPWLRRIAA--------------QHRGDVLDVGCGDGLLAQRLAPV--
Mvan_4082|M.vanbaalenii_PYR-1 AYHPWLVGIAG--------------EHRGDVLDIGCGDGLLAQRLLPV--
MAB_1294c|M.abscessus_ATCC_199 HYQREVLRRVP--------------AHATDVLDVGCGIGMLTRELAPL--
MAV_4317|M.avium_104 YPPEAIDWLLP--------------VGARQVLDLGAGTGKLTTRLVER--
TH_1968|M.thermoresistible__bu YPPEAIDWLLP--------------SDARDVLDLGAGTGKLTTRLVER--
MLBr_01723|M.leprae_Br4923 SYDQRCVDYVRHCFDAIVPDELFTELPYDCALELGCGTGFFLLNLIQSGV
Mflv_4035|M.gilvum_PYR-GCK RETGRLAADLT------------AVSATDHVLDIGCGPGNACRIAAAR--
.*::*.* *
Mb0092|M.bovis_AF2122/97 IPYVTAVDIDAPVLRRAQT-RFA-NAPIRWLHADIMTAELPNAGFDAVVS
Rv0089|M.tuberculosis_H37Rv IPYVTAVDIDAPVLRRAQT-RFA-NAPIRWLHADIMTAELPNAGFDAVVS
MMAR_0244|M.marinum_M VPDVTALDVDADVLRRAER-RFA-DTSIRWVHGDVMTGELPHRGFDAVLA
MUL_4869|M.ulcerans_Agy99 VPDVTALDVDADVLRRAEH-RFA-DTSIWWVHGDVMTGELPHRGFDAVLA
MSMEG_4708|M.smegmatis_MC2_155 SRSVTAIEPDPQTLQRAWV-RLAHLPNVTVAQNDFDTFDPGTHRFDVVTM
Mvan_4082|M.vanbaalenii_PYR-1 SRSVTLIDPDPDSVARAQA-RLAGHADVSISQQSFHDYRPGTALFDVITF
MAB_1294c|M.abscessus_ATCC_199 AQRVVGIDPHEPSLRIARA-ETS-ASNVEYVRGSFLEHPFPAESFDLIAA
MAV_4317|M.avium_104 GLDVVAVDPIPDMLEVLRS-SLP---ETRALLGTAEEIPLEDNSVDVVLV
TH_1968|M.thermoresistible__bu GLNVTAVDPIPEMLELLSK-SLP---DTPALLGTAEEIPLPDNSVDAVLV
MLBr_01723|M.leprae_Br4923 ARRGSVTDLSPGMVKIATRNGQSLGLDINGRVADAEGIPYDDNTFDLVVG
Mflv_4035|M.gilvum_PYR-GCK GARVTGVDPSAAMLRVARIVTHG--ASVTWVRGGAEALPVPDAGATVAWA
: :
Mb0092|M.bovis_AF2122/97 NAALHHIEDTRTALSRLGGLVTPGGTLAVVTFVT--------------PS
Rv0089|M.tuberculosis_H37Rv NAALHHIEDTRTALSRLGGLVTPGGTLAVVTFVT--------------PS
MMAR_0244|M.marinum_M NASLHHIEDTRGALIRLAELVLPAGTLAVVTFVR--------------PS
MUL_4869|M.ulcerans_Agy99 SASLHHIEGTRGALIRLAELVLPGGTLAVVTFVR--------------PS
MSMEG_4708|M.smegmatis_MC2_155 VATLHHMD-LRAALSKARELLTPTGEIAVVGLSAN-------------KT
Mvan_4082|M.vanbaalenii_PYR-1 VASLHHMA-LRESLRKARALLRPGGELAVVGLSAN-------------RS
MAB_1294c|M.abscessus_ATCC_199 VAALHHMD-IEQGLRRMSDLLKPGGRIVIVGFAAN-------------RS
MAV_4317|M.avium_104 AQAWHWVD-PERAIPEVARVLRPGGRLGLVWNTRDERLGWVRELGRIIGS
TH_1968|M.thermoresistible__bu AQAWHWFD-VDRAVREVARVLRPGGRLGLVWNTRDERMGWVKELGRIIGD
MLBr_01723|M.leprae_Br4923 HAVLHHIPDVELSLREVLRVLKPGGRFVFAGEPTDVG-----NRYARVFA
Mflv_4035|M.gilvum_PYR-GCK LATVHHWPDVDAGLAEIHRALGPGARFLAVERQTEP--------------
* .: . : * . : .
Mb0092|M.bovis_AF2122/97 LRNGLWHLTSWVACGMANRVKGKWEHSAPIKWPPPQTLHELRSHVRALLP
Rv0089|M.tuberculosis_H37Rv LRNGLWHLTSWVACGMANRVKGKWEHSAPIKWPPPQTLHELRSHVRALLP
MMAR_0244|M.marinum_M LRNGLWHLTAWLATGVANRVRRKWEHTAAIKWPPPVTFSQLRELVRELLP
MUL_4869|M.ulcerans_Agy99 PRNVLWHLTAWLATGVANRVRRKWDHTAAIKWPPPVTFSQLRELVRELLP
MSMEG_4708|M.smegmatis_MC2_155 LRDWAWSGLCLPAVRLGSFCHRETRDIGVPVAEPAENLDEIRRVAADVLP
Mvan_4082|M.vanbaalenii_PYR-1 ASDWIWSGLCLPAVRIGSRLHRETRDIGVVVAQPRESLREIRLVSDDVIP
MAB_1294c|M.abscessus_ATCC_199 VTDLWYDGLGFWLHRYYRLRYGWWEHPSPVSCEGLDSHDEVRRIAQRVLP
MAV_4317|M.avium_104 DGDGR-THVALPEP-FTDLAHHEVEWTNYLTPQALIDLVASRSYCITSPT
TH_1968|M.thermoresistible__bu EHDPLSKTVTLPEP-FTDVQRHQVEWTNYLTPQALIDLVASRSYCITSPA
MLBr_01723|M.leprae_Br4923 DLTWKVTIRVVQLPGLSAWRRTQVEIDENSRAAALEWVVDLHTFEPKVLE
Mflv_4035|M.gilvum_PYR-GCK ------DATGISSHGWTGAQAETFAAMCESAGLTTVRVGSGRAGKRRVWT
:
Mb0092|M.bovis_AF2122/97 GACIRR---------LLYGRVLVTWRAPV---------------------
Rv0089|M.tuberculosis_H37Rv GACIRR---------LLYGRVLVTWRAPV---------------------
MMAR_0244|M.marinum_M GARVRR---------LLYGRVLITWQAPG---------------------
MUL_4869|M.ulcerans_Agy99 GARVRR---------LLYGRVLITWQAPG---------------------
MSMEG_4708|M.smegmatis_MC2_155 GAMIRR---------ALYYRYLLRWQPAN---------------------
Mvan_4082|M.vanbaalenii_PYR-1 GAVVRR---------ALYYRYLLRWRKPA---------------------
MAB_1294c|M.abscessus_ATCC_199 GARLQR---------LVLWRHLLTWEKS----------------------
MAV_4317|M.avium_104 EVRTRT---------LDQVRHLLATHPALANSTGLALPYVTRCTRATLAG
TH_1968|M.thermoresistible__bu KVRART---------LERVRELLATHPALANNSGLALPYITVCIRATLA-
MLBr_01723|M.leprae_Br4923 TMATNAGAVQVKTVTEEFTAAMLGWPLRTFESTMPPSKLGWGWARFAFTS
Mflv_4035|M.gilvum_PYR-GCK VHATRP--------------------------------------------
.
Mb0092|M.bovis_AF2122/97 --------------------------------
Rv0089|M.tuberculosis_H37Rv --------------------------------
MMAR_0244|M.marinum_M --------------------------------
MUL_4869|M.ulcerans_Agy99 --------------------------------
MSMEG_4708|M.smegmatis_MC2_155 --------------------------------
Mvan_4082|M.vanbaalenii_PYR-1 --------------------------------
MAB_1294c|M.abscessus_ATCC_199 --------------------------------
MAV_4317|M.avium_104 --------------------------------
TH_1968|M.thermoresistible__bu --------------------------------
MLBr_01723|M.leprae_Br4923 WKTLSWVDANVWRRVIPKSWFYNIIITGVKPS
Mflv_4035|M.gilvum_PYR-GCK --------------------------------