For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPDEVVVVIGVGGMGRVIARRQGIGKALLLADHNEQALQAVVDELKADGHNVIGQRVDVSSRASVSGLAQ AAAGLGRVAQVAHTAGLSPAQSPTAAILAVDLVGVALVVEEFGKIVASGGAGVVIASMAGHMPYELTAEQ EQELGTTHAEDLLSLPYVRDIEHPAVAYQVSKRANHIRVRFAARQWGTRGARINSISPGVVSTPMGHQEL ASETGAITKAMVDGSASRRIGTSDDIAAAAAFLLGPESSYVTGTDLLVDGGVIAAVRSGQLFN
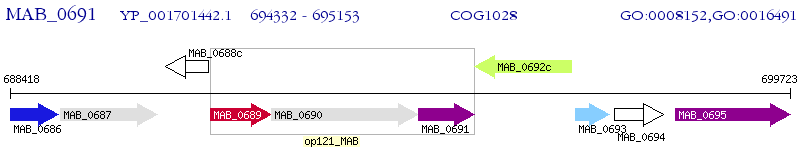
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0691 | - | - | 100% (273) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0827 | - | 4e-13 | 26.74% (258) | putative short-chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_4680c | - | 2e-12 | 25.19% (258) | putative short-chain dehydrogenase/reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0950c | - | 6e-13 | 26.46% (291) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1024 | - | 2e-29 | 33.84% (263) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv0927c | - | 6e-13 | 26.46% (291) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 1e-07 | 25.00% (248) | 3-oxoacyl- |
| M. marinum M | MMAR_0243 | - | 9e-84 | 60.30% (267) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_5240 | - | 2e-31 | 34.08% (267) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6662 | - | 1e-29 | 33.21% (268) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_0784 | - | 2e-29 | 34.36% (259) | short chain dehydrogenase |
| M. ulcerans Agy99 | MUL_4870 | - | 2e-81 | 59.47% (264) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_5812 | - | 2e-28 | 33.46% (263) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0243|M.marinum_M --------MSRDVLTVIGVGGMGQAIARRLGSG-KTVLLADNNADTLASV
MUL_4870|M.ulcerans_Agy99 --------MSRNVLTVIGVGGMGQAIARRLGSG-KTVVLADNNADTLASV
MAB_0691|M.abscessus_ATCC_1997 --------MPDEVVVVIGVGGMGRVIARRQGIG-KALLLADHNEQALQAV
MSMEG_6662|M.smegmatis_MC2_155 --------MRTVSVITGGAGGMGVATAKIVGKE-HAVVLCDVRQDRLDTA
TH_0784|M.thermoresistible__bu ---------------------MGQATARVVGRE-HAVVLCDVRQDRLDTA
MAV_5240|M.avium_104 -----------MSVITGGAGGMGVATAKVVGRD-HTLVLCDVRQDRLTAA
Mvan_5812|M.vanbaalenii_PYR-1 --------MTAVSLITGGAGGMGLATAKVVGQD-RAVVLCDVRQDRLDGA
Mflv_1024|M.gilvum_PYR-GCK --------MSAVSLITGGAGGMGLATAKVVGRD-HSVVLCDVRQERLDAA
Mb0950c|M.bovis_AF2122/97 MILDMFRLDDKVAVITGGGRGLGAAIALAFAQAGADVLIASRTSSELDAV
Rv0927c|M.tuberculosis_H37Rv MILDMFRLDDKVAVITGGGRGLGAAIALAFAQAGADVLIASRTSSELDAV
MLBr_01807|M.leprae_Br4923 --------MTDAAAKVSAAPGRGKPAFVS-----RSILVTGGNRGIGLAV
* ::: . .
MMAR_0243|M.marinum_M SETLAAEGHDVKSRGVDVCAAESVHDLA---QYAATLGAVTQLAHTAGLS
MUL_4870|M.ulcerans_Agy99 AESLAAEGHDVRSRGVDVCAAESVHDLA---QYAATLGAVTQLAHTAGLS
MAB_0691|M.abscessus_ATCC_1997 VDELKADGHNVIGQRVDVSSRASVSGLA---QAAAGLGRVAQVAHTAGLS
MSMEG_6662|M.smegmatis_MC2_155 ATALAEVGITPTVVKCDVTDRHAVNRLF---ATTAGLGTLASVIHTAGVS
TH_0784|M.thermoresistible__bu AAVLRDDGITATPVHCDVTDPQAVDRLF---KETAGIGELASVIHTAGVS
MAV_5240|M.avium_104 TAALAELGMTPTAVNCDVTDRRAVAQLF---DTAAGLGTVVSVIHTAGVS
Mvan_5812|M.vanbaalenii_PYR-1 AAALKDLGVAVTAVNCDVTDRRAVDALF---ETVSRLGTLVSVIHTAGVS
Mflv_1024|M.gilvum_PYR-GCK AAELAGLGISATTVHCDVTDRDAVGRLF---DGAAVAGTLASVIHTAGVS
Mb0950c|M.bovis_AF2122/97 AEQIRAAGRRAHTVAADLAHPEVTAQLAG--QAVGAFGKLDIVVNNVG--
Rv0927c|M.tuberculosis_H37Rv AEQIRAAGRRAHTVAADLAHPEVTAQLAG--QAVGAFGKLDIVVNNVG--
MLBr_01807|M.leprae_Br4923 ARRLVADGHRVAVTHRASVVPDWFFGVECDVTDNDAIGAAFKAVEEHQGP
: * : * .
MMAR_0243|M.marinum_M PAQASAQAILAVDLLGVALVLQEFGAVIAPGGAGVI-IASMAGHLLPPPS
MUL_4870|M.ulcerans_Agy99 PTQASAQAILAVDLVAVALVLQEFGAVIAPGGAGVI-IASMAGHLLPPPS
MAB_0691|M.abscessus_ATCC_1997 PAQSPTAAILAVDLVGVALVVEEFGKIVASGGAGVV-IASMAGHMPYELT
MSMEG_6662|M.smegmatis_MC2_155 PSMGDAEYVIRTNAIGTATINEAFYAAAPRGSAIVN-VASMAAHLLPEEM
TH_0784|M.thermoresistible__bu PSMGSAEYVMRTNAVGTVHVNEAFLRTAGAGAAIVN-VASMAAHMLPAEM
MAV_5240|M.avium_104 PSMGPADYVMRTNAIGTLNVNEAFYAVAGDGSVIVN-VASMAAHMLPAEI
Mvan_5812|M.vanbaalenii_PYR-1 PSMGDAEYVMRTNALGTLNVDEAFYGAATDGAAIVN-VASMAAYLLPAEL
Mflv_1024|M.gilvum_PYR-GCK PSMGDAEYVMRTNALGTVNVGEQFFARAGEGAALVN-VASMAAHILPAEL
Mb0950c|M.bovis_AF2122/97 ---GTMPNTLLSTSTKDLADAFAFNVGTAHALTVAA-VPLMLEHSGGG--
Rv0927c|M.tuberculosis_H37Rv ---GTMPNTLLSTSTKDLADAFAFNVGTAHALTVAA-VPLMLEHSGGG--
MLBr_01807|M.leprae_Br4923 VEVLVANAGITSDMLIMRMSEEQFQKVIDVNLTGVFRVAKLASRNMIKQR
: * . . :. :
MMAR_0243|M.marinum_M AEQ------ERELAHRPPGQLLELDFVGSIVEPAFAYPFAKQANSIRVRA
MUL_4870|M.ulcerans_Agy99 AEQ------EHDLAHRPPGQLLELDFVGSIVEPAFAYPFAKQANSIRVRA
MAB_0691|M.abscessus_ATCC_1997 AEQ------EQELGTTHAEDLLSLPYVRDIEHPAVAYQVSKRANHIRVRF
MSMEG_6662|M.smegmatis_MC2_155 IPTGLFPQAMQAPEVFLKDMLAACEIVPEEARSGIAYALSKSFVRWYSSS
TH_0784|M.thermoresistible__bu VRTNLFARALTDLDGFLSEMLATCDAFPEEGRSGIAYALSKSFVKWYSAA
MAV_5240|M.avium_104 VPTAQFPLALSDADAFMDAMLAACEIAPEQARSGLAYAVSKSFVKWYSQS
Mvan_5812|M.vanbaalenii_PYR-1 IPTSAFPLALQDTEAFMTEMLAACAIAPEEARSGISYAISKSFVRWYSSS
Mflv_1024|M.gilvum_PYR-GCK IPTAVFPAALQDPDTFMAEMLTACEIAPEEARSGIAYALSKSFVRWYSVA
Mb0950c|M.bovis_AF2122/97 -----------------SVINISSTMGRLAARGFAAYGTAKAALAHYTRL
Rv0927c|M.tuberculosis_H37Rv -----------------SVINISSTMGRLAARGFAAYGTAKAALAHYTRL
MLBr_01807|M.leprae_Br4923 FGR---------------YIFMGSVVGLSGVPGQANYAASKSGLIGMARS
* :*
MMAR_0243|M.marinum_M ASRQWGQREARVNSISPGIISTPMGQQELASPVGDGMRAMIAMSGTGRIG
MUL_4870|M.ulcerans_Agy99 ASRQWGQREARVNSISPDIISTPMGQQELASPVGDGMRAMIAMSGTGRIG
MAB_0691|M.abscessus_ATCC_1997 AARQWGTRGARINSISPGVVSTPMGHQELASETGAITKAMVDGSASRRIG
MSMEG_6662|M.smegmatis_MC2_155 QAERFTGRGLRIVSVSPGSVDTEMGRLEEKAGAG----ALVADAAVPRWG
TH_0784|M.thermoresistible__bu QAERFNARGLRIVSVSPGSIDTEMGRLEEQAGAG----AMVADAAVPRWG
MAV_5240|M.avium_104 QAERFNARGLRIVSVSPGSIDTEMGRLEENAGAG----AMVADAAVPRWG
Mvan_5812|M.vanbaalenii_PYR-1 QAERFNGRGLRIVSVSPGSVDTEMGRLEEQAGAG----AMVADAAVPRWG
Mflv_1024|M.gilvum_PYR-GCK QAERFNGKGLRILSVSPGSIDTEMGRLEEQAGAG----AMVADAAVPRWG
Mb0950c|M.bovis_AF2122/97 AALDLCPR-VRVNAIAPGSILTSA--LEVVAANDELRAPMEQATPLRRLG
Rv0927c|M.tuberculosis_H37Rv AALDLCPR-VRVNAIAPGSILTSA--LEVVAANDELRAPMEQATPLRRLG
MLBr_01807|M.leprae_Br4923 LARELGSRSITANVVAPGFIDTEMTRAMDSKYQE----AALQAIPLGRIG
: : ::*. : * . * *
MMAR_0243|M.marinum_M TPDDIAAAAAFLLGPEATFITGADLLVDGGVVAAIRASR---------
MUL_4870|M.ulcerans_Agy99 TPDDIAAAAAFLLGPEATFITGADLLVDGGVVAAIRANG---------
MAB_0691|M.abscessus_ATCC_1997 TSDDIAAAAAFLLGPESSYVTGTDLLVDGGVIAAVRSGQLFN------
MSMEG_6662|M.smegmatis_MC2_155 TPEEMAELFAFCASDRAGYLTGTDILNDGGVVASMRERARVAAEQQQQ
TH_0784|M.thermoresistible__bu TPEEMAELLAFCASAKASYLTGTDILNDGGVIASMTERARAAAETQ--
MAV_5240|M.avium_104 KPEEMAELLAFCASDKAGYLTGTDILNDGGVIASMTERARVTAAG---
Mvan_5812|M.vanbaalenii_PYR-1 KPEEMAELLAFCAGDKAGYLTGTDILNDGGVVASMTERARLAAASSS-
Mflv_1024|M.gilvum_PYR-GCK TAEEMADLLAFCASRKAGYLTGTDILNDGGVIASMTERTRLASQ----
Mb0950c|M.bovis_AF2122/97 DPVDIAAAAVYLASPAGSFLTGKTLEVDGGLTFPNLDLPIPDL-----
Rv0927c|M.tuberculosis_H37Rv DPVDIAAAAVYLASPAGSFLTGKTLEVDGGLTFPNLDLPIPDL-----
MLBr_01807|M.leprae_Br4923 AVEEIAGAVSFLASEDAGYISGAVIPVDGGLGMGH-------------
::* : . . :::* : ***: