For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKTTAAVLFEAGKPFELMELDLDGPGPGEVLVKYTAAGLCHSDLHLTDGDLPPRFPIVGGHEGSGVIEEV GAGVTRVKPGDHVVCSFIPNCGTCRYCCTGRQNLCDMGATILEGCMPDGSFRFHSQGTDFGAMCMLGTFA ERATVSQHSVVKVDDWLPLETAVLVGCGVPSGWGTAVNAGNLRAGDTAVIYGVGGLGINAVQGATAAGCK YVVVVDPVAFKRETALKFGATHAFADAASAAAKVDELTWGQGADAALILVGTVDDEVVSAATAVIGKGGT VVITGLADPAKLTVHVSGTDLTLHEKTIKGSLFGSCNPQYDIVRLLRLYDAGQLMLDELVTTTYNLEQVN QGYQDLRDGKNIRGVIVH
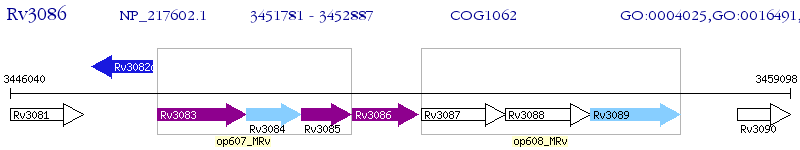
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3086 | adhD | - | 100% (368) | PROBABLE ZINC-TYPE ALCOHOL DEHYDROGENASE ADHD (ALDEHYDE REDUCTASE) |
| M. tuberculosis H37Rv | Rv0761c | adhB | 3e-97 | 47.01% (368) | zinc-containing alcohol dehydrogenase NAD dependent ADHB |
| M. tuberculosis H37Rv | Rv2259 | adhE2 | 3e-54 | 34.72% (360) | zinc-dependent alcohol dehydrogenase AdhE2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3113 | adhD | 0.0 | 100.00% (368) | zinc-type alcohol dehydrogenase AdhD |
| M. gilvum PYR-GCK | Mflv_1486 | - | 1e-110 | 51.63% (368) | alcohol dehydrogenase |
| M. leprae Br4923 | MLBr_01784 | adhE2 | 4e-51 | 32.87% (362) | putative alcohol dehydrogenase (Zn dependent) |
| M. abscessus ATCC 19977 | MAB_2084 | - | 1e-109 | 50.55% (366) | zinc-type alcohol dehydrogenase AdhD |
| M. marinum M | MMAR_0127 | adhD_1 | 0.0 | 86.14% (368) | zinc-containing alcohol dehydrogenase NAD- dependent, |
| M. avium 104 | MAV_2646 | - | 9e-97 | 47.15% (369) | alcohol dehydrogenase B |
| M. smegmatis MC2 155 | MSMEG_3388 | - | 1e-163 | 73.37% (368) | S-(hydroxymethyl)glutathione dehydrogenase/class III alcohol |
| M. thermoresistible (build 8) | TH_2499 | - | 1e-107 | 52.03% (369) | PUTATIVE Alcohol dehydrogenase GroES domain protein |
| M. ulcerans Agy99 | MUL_0470 | adhB | 2e-95 | 45.38% (368) | zinc-containing alcohol dehydrogenase NAD- dependent AdhB |
| M. vanbaalenii PYR-1 | Mvan_0013 | - | 1e-111 | 53.78% (370) | alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv3086|M.tuberculosis_H37Rv -------------------------------------------MKTTAAV
Mb3113|M.bovis_AF2122/97 -------------------------------------------MKTTAAV
MMAR_0127|M.marinum_M -------------------------------------------MKTTAAV
MSMEG_3388|M.smegmatis_MC2_155 MHSDPSHIDLLGPATFTSRDRRPIGFPPLRCSRHKISWKEPSEMKTKAAV
MAB_2084|M.abscessus_ATCC_1997 -------------------------------------------MKTRAAV
Mvan_0013|M.vanbaalenii_PYR-1 -------------------------------------------MKTKAAV
Mflv_1486|M.gilvum_PYR-GCK -------------------------------------------MKTNAAI
TH_2499|M.thermoresistible__bu -------------------------------------------MKMNAAV
MUL_0470|M.ulcerans_Agy99 -------------------------------------------MKTKGAL
MAV_2646|M.avium_104 -------------------------------------------MQTQAAV
MLBr_01784|M.leprae_Br4923 -----------------------------------------MSQTVRGVI
..:
Rv3086|M.tuberculosis_H37Rv LFEAGKPFELMELDLDGPGPGEVLVKYTAAGLCHSDLHLTDGDLPPR-FP
Mb3113|M.bovis_AF2122/97 LFEAGKPFELMELDLDGPGPGEVLVKYTAAGLCHSDLHLTDGDLPPR-FP
MMAR_0127|M.marinum_M LLEAGKPFEIMELELDGPRVGEVLIKYTAAGLCHSDLHLTDGDLPPR-YP
MSMEG_3388|M.smegmatis_MC2_155 SLGVNRPFEIMELELDGPKEGEVLLKNVAAGLCHSDLHLTDGDMPPR-YP
MAB_2084|M.abscessus_ATCC_1997 LWGLEQKWEVEEVDLDPPGPGEVLVRLAATGLCHSDEHLVTGDLPIP-LP
Mvan_0013|M.vanbaalenii_PYR-1 LWGLHQKWEVEELELDAPRAQEVLVKLTASGLCHSDDHLVTGDMPMN-LP
Mflv_1486|M.gilvum_PYR-GCK LWEYGGDWTVEEIDLDPPGDGEVLVSWQATGLCHSDEHIRTGDLPAP-LP
TH_2499|M.thermoresistible__bu LWDVNGDWSIEEVELDGPQEGEVLVSFTATGMCHSDHHVRTGDFPAP-LP
MUL_0470|M.ulcerans_Agy99 IWEFNQPWSVEDIEIGDPVKDEVKIQMEAAGMCRSDNHLVTGGIPMAGFP
MAV_2646|M.avium_104 LFERNTPWSIESIELDAPKATEVLIELHASGMCHSDDHLVTGDMPIR-LP
MLBr_01784|M.leprae_Br4923 SRKKDEPVELVDIVIPDPGPGEAVVDVTACGVCHTDLTYREGGINDR-YP
: .: : * *. : * *:*::* *.: *
Rv3086|M.tuberculosis_H37Rv IVGGHEGSGVIEEVGAGVTRVKPGDHVVCSFIPNCGTCRYCCTGRQNLCD
Mb3113|M.bovis_AF2122/97 IVGGHEGSGVIEEVGAGVTRVKPGDHVVCSFIPNCGTCRYCCTGRQNLCD
MMAR_0127|M.marinum_M IVGGHEGSGVIEEVGPGVSKVKPGDHVVCSFIPNCGTCRYCSTGRQNLCD
MSMEG_3388|M.smegmatis_MC2_155 IVGGHEGSAIVEDVGPGVTKVKPGDHVVCSYIPNCGTCRYCATGRSNLCD
MAB_2084|M.abscessus_ATCC_1997 VVGGHEGAGTVVECGEGVEDLSEGDSVILTFLPSCGRCSYCARGMGNLCE
Mvan_0013|M.vanbaalenii_PYR-1 VVGGHEGAGVVVDVGPGVTEVEAGDHVVLSFIPACGRCTECARGRSNLCV
Mflv_1486|M.gilvum_PYR-GCK LIGGHEGAGIVREVGAGVVGLAPGDHVVASFLPACGRCRFCSTGHQNLCD
TH_2499|M.thermoresistible__bu IIGGHEGAGVVQEVGPGVRDLKPGDHVVASFLPACGRCRWCATGRSNLCD
MUL_0470|M.ulcerans_Agy99 VLGGHEGAGVVTEVGPGVDDIAPGDHVVLSFIPSCGRCPTCQAGLRNLCD
MAV_2646|M.avium_104 CIGGHEGAGVVKAVGEHVSWLSPGDHVVFSFIPSCGRCPSCSTGHQSLCD
MLBr_01784|M.leprae_Br4923 FLLGHEAAGTVEVVGPGVTAVEPGDFVILNWRAVCGQCRACKRGRPSYCF
: ***.:. : * * : ** *: .: . ** * * * . *
Rv3086|M.tuberculosis_H37Rv MGATILEGCMP-DGSFRFH----SQGTDFGAMCMLGTFAERATVSQHSVV
Mb3113|M.bovis_AF2122/97 MGATILEGCMP-DGSFRFH----SQGTDFGAMCMLGTFAERATVSQHSVV
MMAR_0127|M.marinum_M MGATILEGCMS-DGSFRFH----RKGVDFGAMCMLGTFSERATISQHSVV
MSMEG_3388|M.smegmatis_MC2_155 MGATILDGRFP-DGTFRFH----KDGVDYGSLCLLGSFAERTTVSQHSVV
MAB_2084|M.abscessus_ATCC_1997 LGAALMMGPQI-DGTYRFH----ARGEDVGQMCLLGTFSEYTVVPQASVV
Mvan_0013|M.vanbaalenii_PYR-1 YGAAIVAGPQL-DGTFRFHGSSGGRAYDIGQMCVLGTFSEYTVVPIASVV
Mflv_1486|M.gilvum_PYR-GCK LGAMIMAGTQL-DGTYRRHIN----GQDVGVMALVGTFSQYGTVPEASIV
TH_2499|M.thermoresistible__bu LGAMLLAGTQL-DGTYRRHLPD---GRDVGVALLLGTFAQYGVVSEASLV
MUL_0470|M.ulcerans_Agy99 LGAGLLAGAAVSDGTFRIQ----ARGQNVYPMTLLGTFSPYMVVHRSSVV
MAV_2646|M.avium_104 LGAKIYSGMQIHDGTARHHLG----GDDLALACGIGSFAHHTVVHEASCV
MLBr_01784|M.leprae_Br4923 DTFNAQQKMTLIDGTELTP------------ALGIGAFADKTLVHSGQCT
**: :*:*: : . .
Rv3086|M.tuberculosis_H37Rv KVDDWLPLETAVLVGCGVPSGWGTAVNAGNLRAGDTAVIYGVGGLGINAV
Mb3113|M.bovis_AF2122/97 KVDDWLPLETAVLVGCGVPSGWGTAVNAGNLRAGDTAVIYGVGGLGINAV
MMAR_0127|M.marinum_M KIDDWLPLTTAVLVGCGVPTGWGTSVYAGGVRPGDTTVIYGAGGLGINAV
MSMEG_3388|M.smegmatis_MC2_155 KVDEWIPLETAVLVGCGVPAGWGGPVYNNGIRPGDTAVIYGIGGLGINAI
MAB_2084|M.abscessus_ATCC_1997 KIDGGIPLDRAALIGCGVTTGYGSAVRTGEVRAGDTVVVVGAGGIGMNAI
Mvan_0013|M.vanbaalenii_PYR-1 KVDNDIPLDKAALVGCGVTTGYGAAVRTGEAADGDTVVVMGVGGLGINAI
Mflv_1486|M.gilvum_PYR-GCK KIDDDIPLSRACLLGCGVTTGWGSAVNTADVSPGDTVVIIGLGGIGSGAI
TH_2499|M.thermoresistible__bu KIDDDLPLTRACLIGCGVTTGWGSAVNTAKVQPGDTVVVIGFGGVGSGAV
MUL_0470|M.ulcerans_Agy99 KIDPAIPFEVAALVGCGVTTGYGSAVRTADVRPGDDVAIVGLGGVGMAAL
MAV_2646|M.avium_104 KIPDHYRLDLACLLGCGFITGWGSSVYAANVRPGDTVAIAGVGGIGAAAV
MLBr_01784|M.leprae_Br4923 KVDPAADPAVAGLLGCGVMAGLGAAINTAAVSRDDTVAVIGCGGVGDAAI
*: * *:***. :* * .: .* ..: * **:* *:
Rv3086|M.tuberculosis_H37Rv QGATAAGCKYVVVVDPVAFKRETALKFGATHAFADAA-SAAAKVDELTWG
Mb3113|M.bovis_AF2122/97 QGATAAGCKYVVVVDPVAFKRETALKFGATHAFADAA-SAAAKVDELTWG
MMAR_0127|M.marinum_M QGAVSAGAKHVVVVDPVAFKRDTALKFGATHAFPDAA-SAAAKVNEITWG
MSMEG_3388|M.smegmatis_MC2_155 QGAVMAGARYVVAVDPVEFKRDMALKFGATHAFATAE-EAHEAVREITWG
MAB_2084|M.abscessus_ATCC_1997 QGARIAGALAIVAVDPVNFKREQASGFGATHTAASMD-EAWSLVSDMTRG
Mvan_0013|M.vanbaalenii_PYR-1 QGAAIAGARNVVALDPVAYKRERSFEFGATHAAADVA-EAQALITDLTRG
Mflv_1486|M.gilvum_PYR-GCK QGARLAGAEKIVVVEPVETKRDLAFRFGATHFVTSME-EATALVAELTRG
TH_2499|M.thermoresistible__bu QGARLAGAEKIVVVDIVEDKRAKALEFGATHFVTSMA-EATALVGDLTRG
MUL_0470|M.ulcerans_Agy99 QGAVTAGARYIFAVEPVEWKRDQALKFGATHVYPDME-AALMGIAEVTYG
MAV_2646|M.avium_104 QGARLAGARTITVIDPSEYKRAEAVKMGATHTAADWN-EAKAVVAEATWN
MLBr_01784|M.leprae_Br4923 SGAALVGANRIIAVDIDDTKLEWARTFGATHTVNALELEVVKTIQDLTSG
.** .*. : .:: * : :**** . : : * .
Rv3086|M.tuberculosis_H37Rv QGADAALILVGTVDDEVVSAATAVIGKGGTVVITGLADPAKLTVHVSGTD
Mb3113|M.bovis_AF2122/97 QGADAALILVGTVDDEVVSAATAVIGKGGTVVITGLADPAKLTVHVSGTD
MMAR_0127|M.marinum_M QGADQALILVGTVDTEVVSAAAAVIGKGGTVVITGLANPEKLTVQLSGLD
MSMEG_3388|M.smegmatis_MC2_155 QMADGALIFVGTPDEKVVSDAFNVIGKGGTVSLASLADPAKLTVHVSGME
MAB_2084|M.abscessus_ATCC_1997 KLADVCILTTDVAEGSYTAEALALVGKRGRVVVTAIGHPDESSISGSLLE
Mvan_0013|M.vanbaalenii_PYR-1 TMADVCVVTTDSAEGAYVAQALSLVGKRGRVVMTAIPHPTDTVVDMSLFD
Mflv_1486|M.gilvum_PYR-GCK VMADSAILTVGLLAGSMLEEAANIIRKGGAVVMTALSTMADNQPTLGMMM
TH_2499|M.thermoresistible__bu VMADSAILTVGLVEGPMIGECIDIIRKGGAVVLTGIAALADVTPTLPMAM
MUL_0470|M.ulcerans_Agy99 LMAKKVMITVGELQGSDIDTYLAITSKGGTCVVTAIGSLLDTQVNLNLAM
MAV_2646|M.avium_104 RGVDKFICAMGVGDGQLVGQALAMTAKRGRLVVTNIHPMLEREIRANLMD
MLBr_01784|M.leprae_Br4923 FGVDVVIDTVGRPE--TWKQAFYARDLAGTVVLVGVPTP-DMRLDMPLLD
.. : . * :. : .
Rv3086|M.tuberculosis_H37Rv LTLHEKTIKGSLFGSCNPQYDIVRLLRLYDAGQLMLDELVTTTYNLEQVN
Mb3113|M.bovis_AF2122/97 LTLHEKTIKGSLFGSCNPQYDIVRLLRLYDAGQLMLDELVTTTYNLEQVN
MMAR_0127|M.marinum_M LTLNEKTIKGTLFGSANPQYDIVRLLRLYDAGQLKLDELITTQYRLEDVN
MSMEG_3388|M.smegmatis_MC2_155 MTLMEKTIKGCLFGSSNPQYDIVRLLRLYDEGKLKLDELITTRYRLEDIN
MAB_2084|M.abscessus_ATCC_1997 MTLYEKQIRGALYGSSNAPHDIPRLVELYSAGLLKLDELVTREYSLDQIN
Mvan_0013|M.vanbaalenii_PYR-1 LTLYEKQVRGCLFGSSNPRLDIPRLLDLYGSGRLKLDELITREYTLEEIN
Mflv_1486|M.gilvum_PYR-GCK FTLFQKRLLGSLYGEANPRADIPRLLSLYREGKLLLDETVTHEYKLAEVN
TH_2499|M.thermoresistible__bu FTLFQKRLLGSLYGEANPRADIPRLLSLYREGKLLLDETVTHEYKLHDIN
MUL_0470|M.ulcerans_Agy99 LTLMQKNLQGTIFGGGNPQYDIPHLLSMYKARNLNIDDMITRQYRLEQIN
MAV_2646|M.avium_104 LTLTEKQIVGTLYGSGNPRADIPKILELYSAGQVDLESMVTRTYPLEKVN
MLBr_01784|M.leprae_Br4923 FFSHGGSLKSSWYGDCLPERDFPTLVDLYLQGRLPLGKFVSERIGLGDVE
: : . :* . *: :: :* : : . :: * .::
Rv3086|M.tuberculosis_H37Rv QGYQDLRDGKNIRGVIVH--------
Mb3113|M.bovis_AF2122/97 QGYQDLRDGKNIRGVIVH--------
MMAR_0127|M.marinum_M QGYQDLRDGKNIRGVIVHD-------
MSMEG_3388|M.smegmatis_MC2_155 QGYDDLRAGVNIRGVIIHDPEA----
MAB_2084|M.abscessus_ATCC_1997 EGYQDMRSGRNIRGLVRF--------
Mvan_0013|M.vanbaalenii_PYR-1 QGYEDMHNGRNLRGVIRF--------
Mflv_1486|M.gilvum_PYR-GCK EGYEDMREGRNIRGVILHDH------
TH_2499|M.thermoresistible__bu QGYDDMLAGRIIRGVIVHDHS-----
MUL_0470|M.ulcerans_Agy99 EGYQDMIDGKNIRGVIRFTDADR---
MAV_2646|M.avium_104 DGYADMHAGANIRGVLIYPPAEGSLG
MLBr_01784|M.leprae_Br4923 EAFHKIHGGKVLRSVVML--------
:.: .: * :*.::