For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KTTAAVLLEAGKPFEIMELELDGPRVGEVLIKYTAAGLCHSDLHLTDGDLPPRYPIVGGHEGSGVIEEVG PGVSKVKPGDHVVCSFIPNCGTCRYCSTGRQNLCDMGATILEGCMSDGSFRFHRKGVDFGAMCMLGTFSE RATISQHSVVKIDDWLPLTTAVLVGCGVPTGWGTSVYAGGVRPGDTTVIYGAGGLGINAVQGAVSAGAKH VVVVDPVAFKRDTALKFGATHAFPDAASAAAKVNEITWGQGADQALILVGTVDTEVVSAAAAVIGKGGTV VITGLANPEKLTVQLSGLDLTLNEKTIKGTLFGSANPQYDIVRLLRLYDAGQLKLDELITTQYRLEDVNQ GYQDLRDGKNIRGVIVHD*
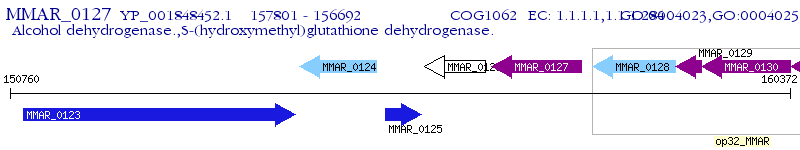
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0127 | adhD_1 | - | 100% (369) | zinc-containing alcohol dehydrogenase NAD- dependent, AdhD_1 |
| M. marinum M | MMAR_3150 | - | e-108 | 50.82% (366) | zinc-containing alcohol dehydrogenase NAD- dependent |
| M. marinum M | MMAR_4935 | adhB | e-101 | 48.64% (368) | zinc-containing alcohol dehydrogenase NAD- dependent AdhB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3113 | adhD | 0.0 | 86.14% (368) | zinc-type alcohol dehydrogenase AdhD |
| M. gilvum PYR-GCK | Mflv_1486 | - | 1e-109 | 51.76% (369) | alcohol dehydrogenase |
| M. tuberculosis H37Rv | Rv3086 | adhD | 0.0 | 86.14% (368) | zinc-type alcohol dehydrogenase AdhD |
| M. leprae Br4923 | MLBr_01784 | adhE2 | 2e-48 | 30.81% (357) | putative alcohol dehydrogenase (Zn dependent) |
| M. abscessus ATCC 19977 | MAB_2084 | - | 1e-106 | 49.73% (366) | zinc-type alcohol dehydrogenase AdhD |
| M. avium 104 | MAV_0705 | - | 1e-103 | 49.18% (368) | alcohol dehydrogenase B |
| M. smegmatis MC2 155 | MSMEG_3388 | - | 1e-167 | 74.80% (369) | S-(hydroxymethyl)glutathione dehydrogenase/class III alcohol |
| M. thermoresistible (build 8) | TH_2499 | - | 1e-109 | 52.97% (370) | PUTATIVE Alcohol dehydrogenase GroES domain protein |
| M. ulcerans Agy99 | MUL_0470 | adhB | 1e-101 | 48.64% (368) | zinc-containing alcohol dehydrogenase NAD- dependent AdhB |
| M. vanbaalenii PYR-1 | Mvan_0013 | - | 1e-112 | 53.51% (370) | alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1486|M.gilvum_PYR-GCK -------------------------------------------MKTNAAI
TH_2499|M.thermoresistible__bu -------------------------------------------MKMNAAV
Mb3113|M.bovis_AF2122/97 -------------------------------------------MKTTAAV
Rv3086|M.tuberculosis_H37Rv -------------------------------------------MKTTAAV
MMAR_0127|M.marinum_M -------------------------------------------MKTTAAV
MSMEG_3388|M.smegmatis_MC2_155 MHSDPSHIDLLGPATFTSRDRRPIGFPPLRCSRHKISWKEPSEMKTKAAV
MAB_2084|M.abscessus_ATCC_1997 -------------------------------------------MKTRAAV
Mvan_0013|M.vanbaalenii_PYR-1 -------------------------------------------MKTKAAV
MAV_0705|M.avium_104 -------------------------------------------MKTKGAL
MUL_0470|M.ulcerans_Agy99 -------------------------------------------MKTKGAL
MLBr_01784|M.leprae_Br4923 -----------------------------------------MSQTVRGVI
. ..:
Mflv_1486|M.gilvum_PYR-GCK LWEYGGDWTVEEIDLDPPGDGEVLVSWQATGLCHSDEHIRTGDLPAP-LP
TH_2499|M.thermoresistible__bu LWDVNGDWSIEEVELDGPQEGEVLVSFTATGMCHSDHHVRTGDFPAP-LP
Mb3113|M.bovis_AF2122/97 LFEAGKPFELMELDLDGPGPGEVLVKYTAAGLCHSDLHLTDGDLPPR-FP
Rv3086|M.tuberculosis_H37Rv LFEAGKPFELMELDLDGPGPGEVLVKYTAAGLCHSDLHLTDGDLPPR-FP
MMAR_0127|M.marinum_M LLEAGKPFEIMELELDGPRVGEVLIKYTAAGLCHSDLHLTDGDLPPR-YP
MSMEG_3388|M.smegmatis_MC2_155 SLGVNRPFEIMELELDGPKEGEVLLKNVAAGLCHSDLHLTDGDMPPR-YP
MAB_2084|M.abscessus_ATCC_1997 LWGLEQKWEVEEVDLDPPGPGEVLVRLAATGLCHSDEHLVTGDLPIP-LP
Mvan_0013|M.vanbaalenii_PYR-1 LWGLHQKWEVEELELDAPRAQEVLVKLTASGLCHSDDHLVTGDMPMN-LP
MAV_0705|M.avium_104 IWEFNQPWSIEEIEIGDPQAHEVKIQMEAAGMCHSDHHLVTGGIPMAGFP
MUL_0470|M.ulcerans_Agy99 IWEFNQPWSVEDIEIGDPVKDEVKIQMEAAGMCRSDNHLVTGGIPMAGFP
MLBr_01784|M.leprae_Br4923 SRKKDEPVELVDIVIPDPGPGEAVVDVTACGVCHTDLTYREGGINDR-YP
: :: : * *. : * *:*::* *.: *
Mflv_1486|M.gilvum_PYR-GCK LIGGHEGAGIVREVGAGVVGLAPGDHVVASFLPACGRCRFCSTGHQNLCD
TH_2499|M.thermoresistible__bu IIGGHEGAGVVQEVGPGVRDLKPGDHVVASFLPACGRCRWCATGRSNLCD
Mb3113|M.bovis_AF2122/97 IVGGHEGSGVIEEVGAGVTRVKPGDHVVCSFIPNCGTCRYCCTGRQNLCD
Rv3086|M.tuberculosis_H37Rv IVGGHEGSGVIEEVGAGVTRVKPGDHVVCSFIPNCGTCRYCCTGRQNLCD
MMAR_0127|M.marinum_M IVGGHEGSGVIEEVGPGVSKVKPGDHVVCSFIPNCGTCRYCSTGRQNLCD
MSMEG_3388|M.smegmatis_MC2_155 IVGGHEGSAIVEDVGPGVTKVKPGDHVVCSYIPNCGTCRYCATGRSNLCD
MAB_2084|M.abscessus_ATCC_1997 VVGGHEGAGTVVECGEGVEDLSEGDSVILTFLPSCGRCSYCARGMGNLCE
Mvan_0013|M.vanbaalenii_PYR-1 VVGGHEGAGVVVDVGPGVTEVEAGDHVVLSFIPACGRCTECARGRSNLCV
MAV_0705|M.avium_104 VLGGHEGAGIVTEVGPGVEDIAPGDHVVLSFIPSCGSCPTCQAGMRNLCD
MUL_0470|M.ulcerans_Agy99 VLGGHEGAGVVTEVGPGVDDIAPGDHVVLSFIPSCGRCPTCQAGLRNLCD
MLBr_01784|M.leprae_Br4923 FLLGHEAAGTVEVVGPGVTAVEPGDFVILNWRAVCGQCRACKRGRPSYCF
.: ***.:. : * ** : ** *: .: . ** * * * . *
Mflv_1486|M.gilvum_PYR-GCK LGAMIMAGTQL-DGTYRRHIN----GQDVGVMALVGTFSQYGTVPEASIV
TH_2499|M.thermoresistible__bu LGAMLLAGTQL-DGTYRRHLPD---GRDVGVALLLGTFAQYGVVSEASLV
Mb3113|M.bovis_AF2122/97 MGATILEGCMP-DGSFRFH----SQGTDFGAMCMLGTFAERATVSQHSVV
Rv3086|M.tuberculosis_H37Rv MGATILEGCMP-DGSFRFH----SQGTDFGAMCMLGTFAERATVSQHSVV
MMAR_0127|M.marinum_M MGATILEGCMS-DGSFRFH----RKGVDFGAMCMLGTFSERATISQHSVV
MSMEG_3388|M.smegmatis_MC2_155 MGATILDGRFP-DGTFRFH----KDGVDYGSLCLLGSFAERTTVSQHSVV
MAB_2084|M.abscessus_ATCC_1997 LGAALMMGPQI-DGTYRFH----ARGEDVGQMCLLGTFSEYTVVPQASVV
Mvan_0013|M.vanbaalenii_PYR-1 YGAAIVAGPQL-DGTFRFHGSSGGRAYDIGQMCVLGTFSEYTVVPIASVV
MAV_0705|M.avium_104 LGAGLLGGEAVSDGTFRIQ----ARGQNVYPMTLLGTFSPYMVVHRSSVV
MUL_0470|M.ulcerans_Agy99 LGAGLLAGAAVSDGTFRIQ----ARGQNVYPMTLLGTFSPYMVVHRSSVV
MLBr_01784|M.leprae_Br4923 DTFNAQQKMTLIDGTELTP------------ALGIGAFADKTLVHSGQCT
**: :*:*: : . .
Mflv_1486|M.gilvum_PYR-GCK KIDDDIPLSRACLLGCGVTTGWGSAVNTADVSPGDTVVIIGLGGIGSGAI
TH_2499|M.thermoresistible__bu KIDDDLPLTRACLIGCGVTTGWGSAVNTAKVQPGDTVVVIGFGGVGSGAV
Mb3113|M.bovis_AF2122/97 KVDDWLPLETAVLVGCGVPSGWGTAVNAGNLRAGDTAVIYGVGGLGINAV
Rv3086|M.tuberculosis_H37Rv KVDDWLPLETAVLVGCGVPSGWGTAVNAGNLRAGDTAVIYGVGGLGINAV
MMAR_0127|M.marinum_M KIDDWLPLTTAVLVGCGVPTGWGTSVYAGGVRPGDTTVIYGAGGLGINAV
MSMEG_3388|M.smegmatis_MC2_155 KVDEWIPLETAVLVGCGVPAGWGGPVYNNGIRPGDTAVIYGIGGLGINAI
MAB_2084|M.abscessus_ATCC_1997 KIDGGIPLDRAALIGCGVTTGYGSAVRTGEVRAGDTVVVVGAGGIGMNAI
Mvan_0013|M.vanbaalenii_PYR-1 KVDNDIPLDKAALVGCGVTTGYGAAVRTGEAADGDTVVVMGVGGLGINAI
MAV_0705|M.avium_104 KIDPSIPFEVAALVGCGVTTGYGSAVRTADIRPGQDVAIVGVGGVGMAAL
MUL_0470|M.ulcerans_Agy99 KIDPAIPFEVAALVGCGVTTGYGSAVRTADVRPGDDVAIVGLGGVGMAAL
MLBr_01784|M.leprae_Br4923 KVDPAADPAVAGLLGCGVMAGLGAAINTAAVSRDDTVAVIGCGGVGDAAI
*:* * *:**** :* * .: .: ..: * **:* *:
Mflv_1486|M.gilvum_PYR-GCK QGARLAGAEKIVVVEPVETKRDLAFRFGATHFVTSMEEATAL-VAELTRG
TH_2499|M.thermoresistible__bu QGARLAGAEKIVVVDIVEDKRAKALEFGATHFVTSMAEATAL-VGDLTRG
Mb3113|M.bovis_AF2122/97 QGATAAGCKYVVVVDPVAFKRETALKFGATHAFADAASAAAK-VDELTWG
Rv3086|M.tuberculosis_H37Rv QGATAAGCKYVVVVDPVAFKRETALKFGATHAFADAASAAAK-VDELTWG
MMAR_0127|M.marinum_M QGAVSAGAKHVVVVDPVAFKRDTALKFGATHAFPDAASAAAK-VNEITWG
MSMEG_3388|M.smegmatis_MC2_155 QGAVMAGARYVVAVDPVEFKRDMALKFGATHAFATAEEAHEA-VREITWG
MAB_2084|M.abscessus_ATCC_1997 QGARIAGALAIVAVDPVNFKREQASGFGATHTAASMDEAWSL-VSDMTRG
Mvan_0013|M.vanbaalenii_PYR-1 QGAAIAGARNVVALDPVAYKRERSFEFGATHAAADVAEAQAL-ITDLTRG
MAV_0705|M.avium_104 QGAVNAGARYIFAIDPVEWKRDQALKFGATHVYPDIMAAMAG-MAEVTYG
MUL_0470|M.ulcerans_Agy99 QGAVTAGARYIFAVEPVEWKRDQALKFGATHVYPDMEAALMG-IAEVTYG
MLBr_01784|M.leprae_Br4923 SGAALVGANRIIAVDIDDTKLEWARTFGATHTVNALELEVVKTIQDLTSG
.** .*. :..:: * : ***** : ::* *
Mflv_1486|M.gilvum_PYR-GCK VMADSAILTVGLLAGSMLEEAANIIRKGGAVVMTALSTMADNQPTLGMMM
TH_2499|M.thermoresistible__bu VMADSAILTVGLVEGPMIGECIDIIRKGGAVVLTGIAALADVTPTLPMAM
Mb3113|M.bovis_AF2122/97 QGADAALILVGTVDDEVVSAATAVIGKGGTVVITGLADPAKLTVHVSGTD
Rv3086|M.tuberculosis_H37Rv QGADAALILVGTVDDEVVSAATAVIGKGGTVVITGLADPAKLTVHVSGTD
MMAR_0127|M.marinum_M QGADQALILVGTVDTEVVSAAAAVIGKGGTVVITGLANPEKLTVQLSGLD
MSMEG_3388|M.smegmatis_MC2_155 QMADGALIFVGTPDEKVVSDAFNVIGKGGTVSLASLADPAKLTVHVSGME
MAB_2084|M.abscessus_ATCC_1997 KLADVCILTTDVAEGSYTAEALALVGKRGRVVVTAIGHPDESSISGSLLE
Mvan_0013|M.vanbaalenii_PYR-1 TMADVCVVTTDSAEGAYVAQALSLVGKRGRVVMTAIPHPTDTVVDMSLFD
MAV_0705|M.avium_104 LMAHKVVVTVGELQGADVDNYLNITQKGGTCVLTAIGSLLDTNVTLNLAM
MUL_0470|M.ulcerans_Agy99 LMAKKVMITVGELQGSDIDTYLAITSKGGTCVVTAIGSLLDTQVNLNLAM
MLBr_01784|M.leprae_Br4923 FGVDVVIDTVGRPE--TWKQAFYARDLAGTVVLVGVPTP-DMRLDMPLLD
.. : .. * :..: .
Mflv_1486|M.gilvum_PYR-GCK FTLFQKRLLGSLYGEANPRADIPRLLSLYREGKLLLDETVTHEYKLAEVN
TH_2499|M.thermoresistible__bu FTLFQKRLLGSLYGEANPRADIPRLLSLYREGKLLLDETVTHEYKLHDIN
Mb3113|M.bovis_AF2122/97 LTLHEKTIKGSLFGSCNPQYDIVRLLRLYDAGQLMLDELVTTTYNLEQVN
Rv3086|M.tuberculosis_H37Rv LTLHEKTIKGSLFGSCNPQYDIVRLLRLYDAGQLMLDELVTTTYNLEQVN
MMAR_0127|M.marinum_M LTLNEKTIKGTLFGSANPQYDIVRLLRLYDAGQLKLDELITTQYRLEDVN
MSMEG_3388|M.smegmatis_MC2_155 MTLMEKTIKGCLFGSSNPQYDIVRLLRLYDEGKLKLDELITTRYRLEDIN
MAB_2084|M.abscessus_ATCC_1997 MTLYEKQIRGALYGSSNAPHDIPRLVELYSAGLLKLDELVTREYSLDQIN
Mvan_0013|M.vanbaalenii_PYR-1 LTLYEKQVRGCLFGSSNPRLDIPRLLDLYGSGRLKLDELITREYTLEEIN
MAV_0705|M.avium_104 LTLMQKNLQGTIFGGGNPQYDIPQLLSMYKAGKLNLDDMITRQYRLEQIN
MUL_0470|M.ulcerans_Agy99 LTLMQKNLQGTIFGGGNPQYDIPHLLSMYKARNLNIDDMITRQYRLEQIN
MLBr_01784|M.leprae_Br4923 FFSHGGSLKSSWYGDCLPERDFPTLVDLYLQGRLPLGKFVSERIGLGDVE
: : . :* . *: *: :* * :.. :: * :::
Mflv_1486|M.gilvum_PYR-GCK EGYEDMREGRNIRGVILHDH---
TH_2499|M.thermoresistible__bu QGYDDMLAGRIIRGVIVHDHS--
Mb3113|M.bovis_AF2122/97 QGYQDLRDGKNIRGVIVH-----
Rv3086|M.tuberculosis_H37Rv QGYQDLRDGKNIRGVIVH-----
MMAR_0127|M.marinum_M QGYQDLRDGKNIRGVIVHD----
MSMEG_3388|M.smegmatis_MC2_155 QGYDDLRAGVNIRGVIIHDPEA-
MAB_2084|M.abscessus_ATCC_1997 EGYQDMRSGRNIRGLVRF-----
Mvan_0013|M.vanbaalenii_PYR-1 QGYEDMHNGRNLRGVIRF-----
MAV_0705|M.avium_104 DGYQDMLDGKNIRGVIRYTDADR
MUL_0470|M.ulcerans_Agy99 EGYQDMIDGKNIRGVIRFTDADR
MLBr_01784|M.leprae_Br4923 EAFHKIHGGKVLRSVVML-----
:.:..: * :*.::