For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RYPLGEALLALYRWRGPLINAGVGGHGYTYLLGAEANRFVFANADAFSWSQTFESLVPVDGPTALIVSDG ADHRRRRSVVAPGLRHHHVQRYVATMVSNIDTVIDGWQPGQRLDIYQELRSAVRRSTAESLFGQRLAVHS DFLGEQLQPLLDLTRRPPQVMRLQQRVNSPGWRRAMAARKRIDDLIDAQIADARTAPRPDDHMLTTLISG CSEEGTTLSDNEIRDSIVSLITAGYETTSGALAWAIYALLTVPGTWESAASEVARVLGGRVPAADDLSAL TYLNGVVHETLRLYSPGVISARRVLRDLWFDGHRIRAGRLLIFSAYVTHRLPEIWPEPTEFRPLRWDPNA ADYRKPAPHEFIPFSGGLHRCIGAVMATTEMTVILARLVARAMLQLPAQRTHRIRAANFAALRPWPGLTV EIRKSAPAQ*
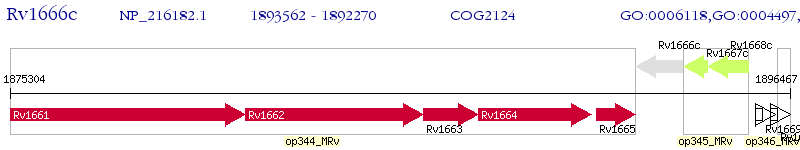
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1666c | cyp139 | - | 100% (430) | Probable cytochrome P450 139 CYP139 |
| M. tuberculosis H37Rv | Rv3685c | cyp137 | 2e-33 | 26.67% (420) | cytochrome P450 137 |
| M. tuberculosis H37Rv | Rv0568 | cyp135B1 | 7e-33 | 26.71% (423) | cytochrome P450 135B1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1694c | cyp139 | 0.0 | 100.00% (430) | cytochrome P450 139 CYP139 |
| M. gilvum PYR-GCK | Mflv_0056 | - | 4e-36 | 30.23% (354) | cytochrome P450 |
| M. leprae Br4923 | MLBr_02088 | - | 3e-17 | 24.21% (347) | putative cytochrome p450 |
| M. abscessus ATCC 19977 | MAB_3423 | - | 1e-29 | 25.70% (393) | cytochrome P450 |
| M. marinum M | MMAR_2475 | cyp139A3 | 0.0 | 79.07% (430) | cytochrome P450 139A3 Cyp139A3 |
| M. avium 104 | MAV_3106 | - | 0.0 | 75.35% (430) | P450 heme-thiolate protein |
| M. smegmatis MC2 155 | MSMEG_0822 | - | 4e-38 | 30.97% (339) | cytochrome P450 |
| M. thermoresistible (build 8) | TH_0136 | - | 8e-37 | 27.92% (419) | PROBABLE CYTOCHROME P450 137 CYP137 |
| M. ulcerans Agy99 | MUL_0473 | cyp51B1 | 7e-30 | 26.16% (367) | cytochrome P450 51B1 Cyp51B1 |
| M. vanbaalenii PYR-1 | Mvan_0245 | - | 2e-40 | 32.30% (322) | cytochrome P450 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0822|M.smegmatis_MC2_155 -----------------------------------MVETASPP-LTELPL
Mvan_0245|M.vanbaalenii_PYR-1 -----------------------------------MVGTVEREDLGALPL
Mflv_0056|M.gilvum_PYR-GCK -----------------------------------MVAGTEPG-VGHLPL
Rv1666c|M.tuberculosis_H37Rv --------------------------------------------------
Mb1694c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_2475|M.marinum_M -------------------------------------------------M
MAV_3106|M.avium_104 --------------------------------------------------
TH_0136|M.thermoresistible__bu ---------------------------------VRPPSLPAPAPLSAVFG
MAB_3423|M.abscessus_ATCC_1997 MATISSTDYLIDQAKRRLPTMNTLPGMGYIENYLNNREWPMTELAAPPPG
MUL_0473|M.ulcerans_Agy99 -----------------------------------MTTAIVPRVSGGEEE
MLBr_02088|M.leprae_Br4923 --------------------------MRTCVPTRTCVYAFIEYLSHNRPM
MSMEG_0822|M.smegmatis_MC2_155 APRNTLPCREQIRCLRSFIDGMQ-------RLRDVGGPVTRIVLGPRWLV
Mvan_0245|M.vanbaalenii_PYR-1 APRNPLPYRQQVRAIRTFHTGLE-------TLRDAGGPVTRLKLAPKWLM
Mflv_0056|M.gilvum_PYR-GCK APKNPLPYRRLLTLVRALDTGQL-------QVRAAGGPITRVQVAPRWLF
Rv1666c|M.tuberculosis_H37Rv -----MRYPLGEALLALYRW---------------RGPLINAGVGG----
Mb1694c|M.bovis_AF2122/97 -----MRYPLGEALLALYRW---------------RGPLINAGVGG----
MMAR_2475|M.marinum_M STYRTVPYRPGEALLALYRR---------------RGAFIDAGVGR----
MAV_3106|M.avium_104 -----MRYRPGEALLALYRR---------------RGPVIDAGAGR----
TH_0136|M.thermoresistible__bu AGYAAAYAVGGNRMVARMLKR--------------YGPVVALPVLG---Y
MAB_3423|M.abscessus_ATCC_1997 SGLKPVMGDQGLPMLGHMVEMFRGGIDWVLNMYQERGPVSWTQTPIG---
MUL_0473|M.ulcerans_Agy99 HGHLEEFRTDPIGLMQRVRD--------------ECGDVGWFQLAN----
MLBr_02088|M.leprae_Br4923 GTNPPSLVEAQMLLLRLIDPGTRADPFPVYRALIDYGPMQLPGMPLT---
: * .
MSMEG_0822|M.smegmatis_MC2_155 PPALLVTSPQGAHDVLSLPDAVADRGGSRNMRQ--LQRLMGGNLLDLPH-
Mvan_0245|M.vanbaalenii_PYR-1 PPMVVATSPQGARDILARGGGHIDK--TRVHHE--MRRLLGANLFDLTH-
Mflv_0056|M.gilvum_PYR-GCK PPFVAVFSPRGVRDVLGRNDAFSER--CIVHEE--VRDMAGDSLFVLPN-
Rv1666c|M.tuberculosis_H37Rv HGYTYLLGAEANRFVFANADAFSWS---QTFES--LVPVDGPTALIVSDG
Mb1694c|M.bovis_AF2122/97 HGYTYLLGAEANRFVFANADAFSWS---QTFES--LVPVDGPTALIVSDG
MMAR_2475|M.marinum_M HGFVYLLGPEANRFVFANADAFSWR---ETFES--LVPVDGPTALIVSDG
MAV_3106|M.avium_104 RGYTLLLGAEANKFVFANADAFSWR---ATFEN--LALVDGPTALIVSDG
TH_0136|M.thermoresistible__bu GTVVAVADPALARQVFAEKPDVLLG--GDGVGP--AAAIYGPRSMFVQEE
MAB_3423|M.abscessus_ATCC_1997 -KIVAALGPDATQAVFSNANKDFSQ---QGWVP--VIGPFFNRGLMLLDF
MUL_0473|M.ulcerans_Agy99 KHVVLLSGAKANEFFFRSSDEELDQ--AEAYPF--MTPIFGKG--VVFDA
MLBr_02088|M.leprae_Br4923 VFSSFSDCDEALRHPLSASDRLKATLAQQAIAAGAEPRPFYASSFMFLDP
. : . .
MSMEG_0822|M.smegmatis_MC2_155 DRWLPRRRTLQPLFTKKHVPRFAGHMAAAAQSVAVSWADG-ATVDLDRAC
Mvan_0245|M.vanbaalenii_PYR-1 EPWLPRRRALQPIFTKQHVREFAGHMATAAQTVADSWADG-TELDLDTEC
Mflv_0056|M.gilvum_PYR-GCK EPWRPRKRALQPVFTPQNVRRFGGHMSRAAEVFVDAWRDG-GDVDLDLEC
Rv1666c|M.tuberculosis_H37Rv ADHRRRRSVVAPGLRHHHVQRYVATMVSNIDTVIDGWQPG-QRLDIYQEL
Mb1694c|M.bovis_AF2122/97 ADHRRRRSVVAPGLRHHHVQRYVATMVSNIDTVIDGWQPG-QRLDIYQEL
MMAR_2475|M.marinum_M EDHRRRRSIVAPGLRHRSVQDYVATMVSTIDNVIDSWRPG-QPLDIYQEF
MAV_3106|M.avium_104 DDHRRRRSVVAPGLRHRQIQDYVTTMVSCIDRVIDGWRPG-QRLDVYQHC
TH_0136|M.thermoresistible__bu PEHLRRRRLLIPPLHGAALAGHVPVIEAATRSAMASWPVG-RPMRMLDAA
MAB_3423|M.abscessus_ATCC_1997 QEHREHRLIMQQAFLRSRLAGYVEQIDAVASEIVAQWPTNDNRFLFYPAI
MUL_0473|M.ulcerans_Agy99 SPERRKEMLHNSALRGEHMKGHATTIEREVHRMIENWGQE-GEIDLLEFF
MLBr_02088|M.leprae_Br4923 PDHTRLRKLVSKAFAPKVVQALEGDIAALVDSLLDKGAAA-GQFDVIADL
. : : : . .
MSMEG_0822|M.smegmatis_MC2_155 RALTLRALGRSVFGVDLDQRAEDVGPALRTSLSWVADRASRPVNLPQWVP
Mvan_0245|M.vanbaalenii_PYR-1 RKLTLRALGRSVLGLDLDEHSDAIAEPLRTALEYIADRALQPISAPRWLP
Mflv_0056|M.gilvum_PYR-GCK RRITMQSLGRSVLGVDLNESADVIAEHMHVASSYTADRALRPVRAPRWLP
Rv1666c|M.tuberculosis_H37Rv RSAVRRSTAESLFGQRLAVHSDFLGEQLQPLLDLTR-RPPQVMRLQQRVN
Mb1694c|M.bovis_AF2122/97 RSAVRRSTAESLFGQRLAVHSDFLGEQLQPLLDLTR-RPPQVMRLQQRVN
MMAR_2475|M.marinum_M RCAVRRSTTESLFGPRLASHSDYLGRQLQPLIDLTH-RLPQVMQLQQRLN
MAV_3106|M.avium_104 RAAVRRSTAESLFGRRLAVHSDALGEYLQPLLDLTH-QPPQLVGLQRRIN
TH_0136|M.thermoresistible__bu RDLTLDVIIRVIFGVHDPAEVARLAAPFETLLALAVSVQTPVRYALRRAG
MAB_3423|M.abscessus_ATCC_1997 KELTLDVASVVFMGNEPHAQHERLEKVNKAFVATTR----AGGAIFRFGL
MUL_0473|M.ulcerans_Agy99 AELTSYTSTSCLIGTKFRNQLDSRFAHFCHELERGT----DPLCYVDPYL
MLBr_02088|M.leprae_Br4923 AFPLAVAVICRLLGVPYEDAPEFGRVSALLVQSVDP-------FITITGE
.:*
MSMEG_0822|M.smegmatis_MC2_155 TR-GQRKARAGNATLHRLAAEILADVRA---DPDRDAPLVRALLEARDPE
Mvan_0245|M.vanbaalenii_PYR-1 TP-ARRRARAASSTLHKLADKILQACRT---DPTRDAPLVQALIAASDPA
Mflv_0056|M.gilvum_PYR-GCK TP-ARRRAAHAVAEMRQVAADMLQVCRD---DPDRDAPLVRALMDARDPE
Rv1666c|M.tuberculosis_H37Rv SP-GWRRAMAARKRIDDLIDAQIADART---APRPDDHMLTTLISGCS-E
Mb1694c|M.bovis_AF2122/97 SP-GWRRAMAARKRIDDLIDAQIADART---APRPDDHMLTTLISGCS-E
MMAR_2475|M.marinum_M SP-GWRRAMAARTRIDELIDAEIANARA---EPSPDDRMLTALINGRS-E
MAV_3106|M.avium_104 AP-AWRRAMAARQRINNLVDTLIAEARA---APNPDDHMLTMLIDGRG-D
TH_0136|M.thermoresistible__bu ALRFWHAYRDANRRIDELVRPLIAARRVP--DAGPRDDILSMLVAART-E
MAB_3423|M.abscessus_ATCC_1997 PPFKWWQGLQGRKLLEEYFAERVGIARQ----KTEGADMLTALCHAET-D
MUL_0473|M.ulcerans_Agy99 PIESFRRRDEARKGLVALVQDIMHQRVANPPTDKRDRDMLDVLVSITDEQ
MLBr_02088|M.leprae_Br4923 PPEATEERLRAGVWLRDYLEQLVKCRRG-----TPGEDLISRLIELDE--
. . : : :: *
MSMEG_0822|M.smegmatis_MC2_155 TGRGLTDDQICDELVLFMLAGHDTTSTTLCYSLWALGHHPDIQERVYAEV
Mvan_0245|M.vanbaalenii_PYR-1 TGRPLSDDEIRDELIVFMLAGHDTTATTLTYAMWALGHHHDVQDRVRAEV
Mflv_0056|M.gilvum_PYR-GCK TGMALSDDDICNDLLIFMLAGHDTTATALTYALWVLGHHPDVQDRVAAEA
Rv1666c|M.tuberculosis_H37Rv EGTTLSDNEIRDSIVSLITAGYETTSGALAWAIYALLTVPGTWESAASEV
Mb1694c|M.bovis_AF2122/97 EGTTLSDNEIRDSIVSLITAGYETTSGALAWAIYALLTVPGTWESAASEV
MMAR_2475|M.marinum_M EGCALSDNEIRDSIVSLIAAGYETTSGALAWATYSLLTLPGAWETAAREV
MAV_3106|M.avium_104 EGYALNDNEIRDAIVSLVTAGYETTSGALAWAVYLLLSQPGAWATAAGEV
TH_0136|M.thermoresistible__bu DGDRLTDDEIRDDLITLVLAGHETTATTLAWLIDYLVHHPAALDRVRAEA
MAB_3423|M.abscessus_ATCC_1997 EGDAFTDTDIVNHMIFLMMAAHDTTTSTTTTMAYYLAANPEWQERVRDES
MUL_0473|M.ulcerans_Agy99 GNPRFCTDEVTGMFISLMFAGHHTSSGTSAWTLIELLRHPDAYAAVIDEL
MLBr_02088|M.leprae_Br4923 SGDQLTEEEIIATCGLLLVAGHETTVNLIANAVLAMLRNPSQWKALSSNP
. : :: :: *.:.*: : :
MSMEG_0822|M.smegmatis_MC2_155 A-ALGD-RELTTDDVPQLTYTMRVLHEALRLCPPGSGTMRMLNEEMTVDG
Mvan_0245|M.vanbaalenii_PYR-1 V-GIGD-RELTPEDVPALGYTIQVLHEALRLCPPAAATARMAMRDVEVDG
Mflv_0056|M.gilvum_PYR-GCK A-ALGR-RELEPGDVPHLKYTTQVLLEALRLCPPAAGVGRMAVRDIDVDG
Rv1666c|M.tuberculosis_H37Rv ARVLGG-RVPAADDLSALTYLNGVVHETLRLYSPGVISARRVLRDLWFDG
Mb1694c|M.bovis_AF2122/97 ARVLGG-RVPAADDLSALTYLNGVVHETLRLYSPGVISARRVLRDLWFDG
MMAR_2475|M.marinum_M ARVLGG-TAPSAETLGALTYLNGVVHETLRLYSPGVVSARRLIRDLWFDG
MAV_3106|M.avium_104 RRVLAG-RLPAAADLSGLTYLNGVVHETLRLYPPGVISARRVMRDLRFEG
TH_0136|M.thermoresistible__bu APGGAQ-RGAGP-------YTEAVINETLRLRPPGPVTGRTTAGHYRLGD
MAB_3423|M.abscessus_ATCC_1997 D-RLGD-GPLDIDSLEKLESLDLVMNEALRLVTPLPFNIRSTVRDTDLLG
MUL_0473|M.ulcerans_Agy99 DELYADGQLVSFHALRQIPRLENVLKETLRLHPPLIILMRVAKGEFQVEG
MLBr_02088|M.leprae_Br4923 Q------------------RAPLVVEETLRYDPAIHLIGRVAAKDMTIGQ
*: *:** .. * . .
MSMEG_0822|M.smegmatis_MC2_155 YRVEAGTVAIVNFYAMHRSPTLWDAPERFDPDRFSPERSAGRN--RWQYL
Mvan_0245|M.vanbaalenii_PYR-1 YRIEAGSMMAVGIYALHRDPALWERPLVFDPDRFSAQNSGGRD--RWQYL
Mflv_0056|M.gilvum_PYR-GCK HRVQAGTLVAVGIYALHRDPEIWPDPDRFDPDRFSPEQTAQRD--RWHFL
Rv1666c|M.tuberculosis_H37Rv HRIRAGRLLIFSAYVTHRLPEIWPEPTEFRPLRWDPNAADYRKPAPHEFI
Mb1694c|M.bovis_AF2122/97 HRIRAGRLLIFSAYVTHRLPEIWPEPTEFRPLRWDPNAADYRKPAPHEFI
MMAR_2475|M.marinum_M HRINSGRLLIFSAYVTHRIPEVWPNPTQFLPQRWDPAAPDYRKPAPHEFI
MAV_3106|M.avium_104 RRIRSGRLLIFSPYVTHRLPEIWPEPMRFAPERWNPDAPGYRRPAPHEFI
TH_0136|M.thermoresistible__bu YTLAPRTRIVLLLGVINRDPNSYPDPHDFRPDRFLHTRP-----DPYAWV
MAB_3423|M.abscessus_ATCC_1997 FHIPAGTMINIWPGMNHRLPELWTEPDKFDPDRFSEPRNEHKR-HRYAFS
MUL_0473|M.ulcerans_Agy99 YPIHEGELVAASPAISNRIAEDFPDPDEFVPERYQEPRQEDLIN-RWTWI
MLBr_02088|M.leprae_Br4923 TTLTEGDTMVLLLAAANRDPAVYSRPDEFDPDRPS-----------SRHL
: :* . : * * * *
MSMEG_0822|M.smegmatis_MC2_155 PFGGGPRSCIGDHFAMLEATLALATVIRAVSVESQNTDLPVETPFTVIAA
Mvan_0245|M.vanbaalenii_PYR-1 PFGAGPRSCIGDHFAMLEATLALATIIRSAEIRSSDPHFPMIVPFTTVAA
Mflv_0056|M.gilvum_PYR-GCK PFAGGARSCIGEHFARLETTLALATIVRSMRVTSSDNDFPVDVPFTTVAH
Rv1666c|M.tuberculosis_H37Rv PFSGGLHRCIGAVMATTEMTVILARLVARAMLQLPAQRTHRIRAANFAAL
Mb1694c|M.bovis_AF2122/97 PFSGGLHRCIGAVMATTEMTVILARLVARAMLQLPAQRTHRIRAANFAAL
MMAR_2475|M.marinum_M PFSGGLHRCIGAVMATTELVVLLARLVARTRLRLPAQR---IRAANFAAL
MAV_3106|M.avium_104 PFSAGLHRCVGAAMATTEMTVMLARLLARTRLRLPAQR---LRAANVAAL
TH_0136|M.thermoresistible__bu PFGGGVKRCLGAAFAMRELTTILRILLTEADLRPVNARPESAAGRAAAVV
MAB_3423|M.abscessus_ATCC_1997 PFGGGAHKCIGMVFGQLEIKAVMHRLLRRYRFELAHPHYQPKWDY-AGMP
MUL_0473|M.ulcerans_Agy99 PFGAGRHRCVGAAFATMQIKAIFSVLLREYEFEMAQPADSYRNDHSKMVV
MLBr_02088|M.leprae_Br4923 AFAVGSHFCLGAALARLEATVTLSAISARFPQVQLAGELVYKPNVAMRGM
.*. * : *:* :. : : :
MSMEG_0822|M.smegmatis_MC2_155 APVPARVTRR------------------
Mvan_0245|M.vanbaalenii_PYR-1 EPIRARVKSVNRIPAADSGGR-------
Mflv_0056|M.gilvum_PYR-GCK GPIPAHLTPR------------------
Rv1666c|M.tuberculosis_H37Rv RPWPGLTVEIRKSAPAQ-----------
Mb1694c|M.bovis_AF2122/97 RPWPGLTVEIRKSAPAQ-----------
MMAR_2475|M.marinum_M APKPGLRVEFVDSVPAQ-----------
MAV_3106|M.avium_104 RPTPGLTVEVIDSVPAQ-----------
TH_0136|M.thermoresistible__bu VPRHGTPVYFRPTAPAASPRCAGRTSTV
MAB_3423|M.abscessus_ATCC_1997 LPIDGMPIILRPLH--------------
MUL_0473|M.ulcerans_Agy99 QLARPARVRYRRRKLSDNRGH-------
MLBr_02088|M.leprae_Br4923 SALPVQV---------------------