For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAGTEPGVGHLPLAPKNPLPYRRLLTLVRALDTGQLQVRAAGGPITRVQVAPRWLFPPFVAVFSPRGVRD VLGRNDAFSERCIVHEEVRDMAGDSLFVLPNEPWRPRKRALQPVFTPQNVRRFGGHMSRAAEVFVDAWRD GGDVDLDLECRRITMQSLGRSVLGVDLNESADVIAEHMHVASSYTADRALRPVRAPRWLPTPARRRAAHA VAEMRQVAADMLQVCRDDPDRDAPLVRALMDARDPETGMALSDDDICNDLLIFMLAGHDTTATALTYALW VLGHHPDVQDRVAAEAAALGRRELEPGDVPHLKYTTQVLLEALRLCPPAAGVGRMAVRDIDVDGHRVQAG TLVAVGIYALHRDPEIWPDPDRFDPDRFSPEQTAQRDRWHFLPFAGGARSCIGEHFARLETTLALATIVR SMRVTSSDNDFPVDVPFTTVAHGPIPAHLTPR*
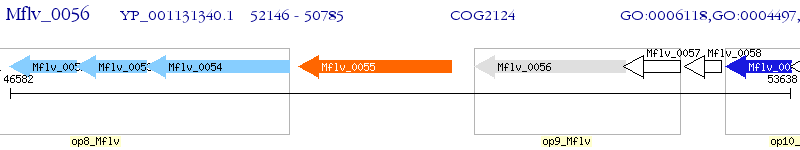
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0056 | - | - | 100% (453) | cytochrome P450 |
| M. gilvum PYR-GCK | Mflv_4801 | - | 1e-41 | 33.90% (351) | cytochrome P450 |
| M. gilvum PYR-GCK | Mflv_1527 | - | 3e-28 | 29.75% (363) | cytochrome P450 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1694c | cyp139 | 3e-36 | 30.23% (354) | cytochrome P450 139 CYP139 |
| M. tuberculosis H37Rv | Rv1666c | cyp139 | 3e-36 | 30.23% (354) | cytochrome P450 139 CYP139 |
| M. leprae Br4923 | MLBr_02088 | - | 8e-15 | 23.24% (327) | putative cytochrome p450 |
| M. abscessus ATCC 19977 | MAB_1426 | - | 2e-37 | 28.50% (393) | putative cytochrome P450 |
| M. marinum M | MMAR_0852 | cyp185A4 | 1e-142 | 54.12% (449) | cytochrome P450 185A4 Cyp185A4 |
| M. avium 104 | MAV_3106 | - | 2e-37 | 28.85% (357) | P450 heme-thiolate protein |
| M. smegmatis MC2 155 | MSMEG_0968 | - | 0.0 | 74.62% (457) | cytochrome P450 |
| M. thermoresistible (build 8) | TH_3255 | - | 1e-178 | 66.89% (453) | PUTATIVE cytochrome P450 |
| M. ulcerans Agy99 | MUL_0604 | cyp185A4 | 1e-138 | 52.78% (449) | cytochrome P450 185A4 Cyp185A4 |
| M. vanbaalenii PYR-1 | Mvan_0858 | - | 0.0 | 70.92% (447) | cytochrome P450 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1694c|M.bovis_AF2122/97 -----------------------------------------------MRY
Rv1666c|M.tuberculosis_H37Rv -----------------------------------------------MRY
MAV_3106|M.avium_104 -----------------------------------------------MRY
Mflv_0056|M.gilvum_PYR-GCK ---------MV------------------AGTEPGVGHLPLAPKNPLPYR
MSMEG_0968|M.smegmatis_MC2_155 ---------MVSDVPAVTAPPGRSADASGANAKPDTGALPLAPRNPLSLR
Mvan_0858|M.vanbaalenii_PYR-1 ---------MTAR---------------DTAAAPEVGSLPLVPRNPLPLW
TH_3255|M.thermoresistible__bu ---------MVVT-----------------TERADVATLPPAPRNPLPYW
MMAR_0852|M.marinum_M ---------MVDS--------------------PDVRSLPLAPRNPLPYW
MUL_0604|M.ulcerans_Agy99 ---------MVDP--------------------PDIRSLPLAPRNPLPYW
MAB_1426|M.abscessus_ATCC_1997 -MSASALQPEPLSAPVEVAAGECPVNHDLYPGGPTFLRWHHTRVGYLKSW
MLBr_02088|M.leprae_Br4923 MRTCVPTR------------------TCVYAFIEYLSHNRPMGTNPPSLV
Mb1694c|M.bovis_AF2122/97 PLGE-ALLALYRWRGPLINAG-----------VGGHGYTYLLGAEANRFV
Rv1666c|M.tuberculosis_H37Rv PLGE-ALLALYRWRGPLINAG-----------VGGHGYTYLLGAEANRFV
MAV_3106|M.avium_104 RPGE-ALLALYRRRGPVIDAG-----------AGRRGYTLLLGAEANKFV
Mflv_0056|M.gilvum_PYR-GCK RLLT-LVRALDTGQLQVRAAGGPITRVQVAPRWLFPPFVAVFSPRGVRDV
MSMEG_0968|M.smegmatis_MC2_155 QMAR-AVRALDMGQQVIRAAGGAVTRVQFGPRWLVPPLVAVFSPEGIRDV
Mvan_0858|M.vanbaalenii_PYR-1 KLVQ-LVRRLDTGQEVIRDAGGPITRIQLGPKKLMPPIVAVMSPAGMRDV
TH_3255|M.thermoresistible__bu RQVR-AVRSFHTGQELLRDAGGPVK-------WAIPPIVLVTSPAGIRDV
MMAR_0852|M.marinum_M QQLK-ALKTLHVGSEALRDAGGSVTRIWLGPKRLVPPMVMVTSPQGARDV
MUL_0604|M.ulcerans_Agy99 QQLK-ALKTLHVGSEALRDAGGSVTRIWLGPKRLVPPMVMVTSPQGARDV
MAB_1426|M.abscessus_ATCC_1997 RLALGAVWSTYRHTISEYLADLPGQDDVIVARAPMRKAVIVRNPELARHV
MLBr_02088|M.leprae_Br4923 EAQMLLLRLIDPGTRADPFPVYRALIDYGPMQLPGMPLTVFSSFSDCDEA
: . . . . .
Mb1694c|M.bovis_AF2122/97 FANADAFSWS---QTFESLVPVDGP------TALIVSDGADHRRRRSVVA
Rv1666c|M.tuberculosis_H37Rv FANADAFSWS---QTFESLVPVDGP------TALIVSDGADHRRRRSVVA
MAV_3106|M.avium_104 FANADAFSWR---ATFENLALVDGP------TALIVSDGDDHRRRRSVVA
Mflv_0056|M.gilvum_PYR-GCK LGRNDAFSER--CIVHEEVRDMAGD------SLFVLPN-EPWRPRKRALQ
MSMEG_0968|M.smegmatis_MC2_155 LGRNDAFAER--CIVHDEVRHAAGD------SLFVLPN-EQWRPRKRALQ
Mvan_0858|M.vanbaalenii_PYR-1 LGRNDASSDR--CIIHEQVQEMAGD------SLWVLPN-EQWRPRKRALA
TH_3255|M.thermoresistible__bu LNRRDALSER--CQVHHEVRHLGGD------SLFVLPT-DPWRARRRALA
MMAR_0852|M.marinum_M LGRSGALSDRGTLPLLKDLRRAIGE------SVLNLPH-DTWLPRRRTLQ
MUL_0604|M.ulcerans_Agy99 LGRSGALSDRGTLPLLKDLRRAIGE------SVLNLPH-DTWLPRRRTLQ
MAB_1426|M.abscessus_ATCC_1997 LVANQDNYIKS--AEYDLLAVGFGR------GLVTDLNEGLWNRNRRLVQ
MLBr_02088|M.leprae_Br4923 LRHPLSASDRLKATLAQQAIAAGAEPRPFYASSFMFLDPPDHTRLRKLVS
: . . : :
Mb1694c|M.bovis_AF2122/97 PGLRHHHVQRYVATMVSNIDTVIDGWQ------PGQRLDIYQELRSAVRR
Rv1666c|M.tuberculosis_H37Rv PGLRHHHVQRYVATMVSNIDTVIDGWQ------PGQRLDIYQELRSAVRR
MAV_3106|M.avium_104 PGLRHRQIQDYVTTMVSCIDRVIDGWR------PGQRLDVYQHCRAAVRR
Mflv_0056|M.gilvum_PYR-GCK PVFTPQNVRRFGGHMSRAAEVFVDAWR------DGGDVDLDLECRRITMQ
MSMEG_0968|M.smegmatis_MC2_155 PVFTKHSVRAFGGHMSRAAQTLVDEWADQWAQADSLEVDLDAVCRRVTMQ
Mvan_0858|M.vanbaalenii_PYR-1 PVFTKTSVRAFGGHMSKAAQAFVDRWP------SGGEVDLDAECRRVAMQ
TH_3255|M.thermoresistible__bu PVFTPTAVRSYGGYMSEAAQRVADGWQ------AAREVDLDAECRRLTMR
MMAR_0852|M.marinum_M PIFTKQRVAGFAGHMSEAANHILLGWP------DGAEVDLNAASHALVLR
MUL_0604|M.ulcerans_Agy99 PIFTKQRVAGFAGHMSEAANHILLGWP------DGAEVDLNAASHALVLR
MAB_1426|M.abscessus_ATCC_1997 PIFAKRQVDLFAPQMAEAAARTISRWD--ELYAEGKPVDITAEMNYLTMD
MLBr_02088|M.leprae_Br4923 KAFAPKVVQALEGDIAALVDSLLDKGA------AAGQFDVIADLAFPLAV
: : : . .*:
Mb1694c|M.bovis_AF2122/97 STAESLFGQRLA-----------------------------VHSDFLGEQ
Rv1666c|M.tuberculosis_H37Rv STAESLFGQRLA-----------------------------VHSDFLGEQ
MAV_3106|M.avium_104 STAESLFGRRLA-----------------------------VHSDALGEY
Mflv_0056|M.gilvum_PYR-GCK SLGRSVLGVDLN-----------------------------ESADVIAEH
MSMEG_0968|M.smegmatis_MC2_155 SLGRSVLGIDLN-----------------------------ERADVIVEH
Mvan_0858|M.vanbaalenii_PYR-1 SLGRSVLGVDLN-----------------------------ERGETIARC
TH_3255|M.thermoresistible__bu SLGRSVLGLDIN-----------------------------ERADAISEP
MMAR_0852|M.marinum_M VVGHSVFGLDLQ-----------------------------DKADVITTG
MUL_0604|M.ulcerans_Agy99 VVGHSVFGLDLQ-----------------------------DKADVITTG
MAB_1426|M.abscessus_ATCC_1997 IVAQTMFGIDLSGDMAERMRIYFARLLKLFGVGFIVGAAPPLRWVVDKLA
MLBr_02088|M.leprae_Br4923 AVICRLLGVPYE-----------------------------------DAP
::*
Mb1694c|M.bovis_AF2122/97 LQPLLDLTR-RPPQVMRLQQRVNSPGWRRAMAARKRIDDLIDAQIADART
Rv1666c|M.tuberculosis_H37Rv LQPLLDLTR-RPPQVMRLQQRVNSPGWRRAMAARKRIDDLIDAQIADART
MAV_3106|M.avium_104 LQPLLDLTH-QPPQLVGLQRRINAPAWRRAMAARQRINNLVDTLIAEARA
Mflv_0056|M.gilvum_PYR-GCK MHVASSYTADRALRPVRAPRWLPTPARRRAAHAVAEMRQVAADMLQVCRD
MSMEG_0968|M.smegmatis_MC2_155 MHVASSYATDRALRPLRAPRWLPTPARRRARAAVAAMRAVTAEILQACRD
Mvan_0858|M.vanbaalenii_PYR-1 MHVASSYTADRALRPVRAPRWLLTPARQRARSAVNAMRAITDEMVKACRD
TH_3255|M.thermoresistible__bu MHIASSYTADRALRPVRAPRWLPTPARRRARAAVATMRRITDEMLQACRA
MMAR_0852|M.marinum_M VTLAAKWAADRSARPVRAPHWLPTPARRRARGAYDAMYKIAADMLAACRA
MUL_0604|M.ulcerans_Agy99 VTLAAKWAADRSARPVRAPHWLPTPARPRARGAYDAMYKIAADMLAACRA
MAB_1426|M.abscessus_ATCC_1997 AHGPDELSSHTPRLAIRALRIGASVAAPRTMKGLRWVERTIDQLIADHRS
MLBr_02088|M.leprae_Br4923 EFGRVSALLVQSVDPFITITGEPPEATEERLRAGVWLRDYLEQLVKCRRG
. . . . . . . : : *
Mb1694c|M.bovis_AF2122/97 A-PRPDDHMLTTLISGCS-EEGTTLSDNEIRDSIVSLITAGYETTSGALA
Rv1666c|M.tuberculosis_H37Rv A-PRPDDHMLTTLISGCS-EEGTTLSDNEIRDSIVSLITAGYETTSGALA
MAV_3106|M.avium_104 A-PNPDDHMLTMLIDGRG-DEGYALNDNEIRDAIVSLVTAGYETTSGALA
Mflv_0056|M.gilvum_PYR-GCK D-PDRDAPLVRALMDARDPETGMALSDDDICNDLLIFMLAGHDTTATALT
MSMEG_0968|M.smegmatis_MC2_155 D-ADRDAPLVRALMAAEDPETGERLSDEDICNDLLIFMLAGHDTTATALT
Mvan_0858|M.vanbaalenii_PYR-1 D-PTRDAPLVQALIAATDPETGRPLTDEEISNDLLIFMLAGHDTTATALT
TH_3255|M.thermoresistible__bu D-PDRDAPLVHALIRATDPETGRGLSDEDICNDLLIFMLAGHDTTATALT
MMAR_0852|M.marinum_M D-PNREAPLIRALIDATDPNTGESLSDQQICDEMVYFMILGEDTTSTVLT
MUL_0604|M.ulcerans_Agy99 D-PSREAPLITALIDATGPNTGESLSDQQICDEMVYFMILGEDTTSTVLT
MAB_1426|M.abscessus_ATCC_1997 GRIARQDNLLALLMAAEDPETGAKYTDLEIRDELMTFLGAGFETTAAALA
MLBr_02088|M.leprae_Br4923 T---PGEDLISRLIELDE--SGDQLTEEEIIATCGLLLVAGHETTVNLIA
:: *: * .: :* :: * :** ::
Mb1694c|M.bovis_AF2122/97 WAIYALLTVPGTWESAASEVARVLGGRVPAADDLSALTYLNGVVHETLRL
Rv1666c|M.tuberculosis_H37Rv WAIYALLTVPGTWESAASEVARVLGGRVPAADDLSALTYLNGVVHETLRL
MAV_3106|M.avium_104 WAVYLLLSQPGAWATAAGEVRRVLAGRLPAAADLSGLTYLNGVVHETLRL
Mflv_0056|M.gilvum_PYR-GCK YALWVLGHHPDVQDRVAAEAA-ALGRRELEPGDVPHLKYTTQVLLEALRL
MSMEG_0968|M.smegmatis_MC2_155 YALWALGHHPEIQDRVAAEAR-ALGDRELTPADVPQLGYTVQVLHESLRM
Mvan_0858|M.vanbaalenii_PYR-1 HALWVLGHHPDIQDRVAAEAA-AIGDRELTPDDVPALGYTVQVLHESVRL
TH_3255|M.thermoresistible__bu YALWALGHHPEIQDRVAAEVA-ELPDREITPHDVPRLGYTVQVLRESLRL
MMAR_0852|M.marinum_M YALWALGRHSQLQDRVAAEAA-ELGDRPLTPADVPRLGYTIQVLQEAMRL
MUL_0604|M.ulcerans_Agy99 YALWALGQHCQLQDRLAAEAA-ELGDRPLTPADVPRLGYTIQVLQEAMRL
MAB_1426|M.abscessus_ATCC_1997 WTWYLLSRNPDARAKLGQEVDRVLGGRQPTAADVDNLPWTAAVLNEAMRV
MLBr_02088|M.leprae_Br4923 NAVLAMLRNPSQWKALSSNPQ-----------------RAPLVVEETLRY
: : . : *: *::*
Mb1694c|M.bovis_AF2122/97 YSPGVISARRVLRDLWFDGHRIRAGRLLIFSAYVTHRLPEIWPEPTEFRP
Rv1666c|M.tuberculosis_H37Rv YSPGVISARRVLRDLWFDGHRIRAGRLLIFSAYVTHRLPEIWPEPTEFRP
MAV_3106|M.avium_104 YPPGVISARRVMRDLRFEGRRIRSGRLLIFSPYVTHRLPEIWPEPMRFAP
Mflv_0056|M.gilvum_PYR-GCK CPPAAGVGRMAVRDIDVDGHRVQAGTLVAVGIYALHRDPEIWPDPDRFDP
MSMEG_0968|M.smegmatis_MC2_155 CPPAAGVGRLALRDIAVAGHRVEAGSLVALGIYAVHHDPQLWPDPEVFDP
Mvan_0858|M.vanbaalenii_PYR-1 CPPAAGVARLVTRDFAVDGYRVEAGSLVALGLYALHHDPALWPDPMAFDP
TH_3255|M.thermoresistible__bu CPPAAGVGRLAMSDLAVDGYRVEAGTLLAVGIQAIHRDPSLWPDPMVFDP
MMAR_0852|M.marinum_M CPPGPTVARRAMQDIEVDGYRVQAGTYCLVGVYAMHRDPALWDDPLAFDP
MUL_0604|M.ulcerans_Agy99 RPPGPTVARRAMQDIEVDGYRVQAGTYCLVGVYAMHRDPALWDDPLAFDP
MAB_1426|M.abscessus_ATCC_1997 YPPILGLARTAKADDVLGDYPISAGTTVMVLIDSIHHNERVWDDAKTFDP
MLBr_02088|M.leprae_Br4923 DPAIHLIGRVAAKDMTIGQTTLTEGDTMVLLLAAANRDPAVYSRPDEFDP
.. .* . * . : * . :: :: . * *
Mb1694c|M.bovis_AF2122/97 LRWDPNAADYRKPAPHEFIPFSGGLHRCIGAVMATTEMTVILARLVARAM
Rv1666c|M.tuberculosis_H37Rv LRWDPNAADYRKPAPHEFIPFSGGLHRCIGAVMATTEMTVILARLVARAM
MAV_3106|M.avium_104 ERWNPDAPGYRRPAPHEFIPFSAGLHRCVGAAMATTEMTVMLARLLARTR
Mflv_0056|M.gilvum_PYR-GCK DRFSPEQTAQRD--RWHFLPFAGGARSCIGEHFARLETTLALATIVRSMR
MSMEG_0968|M.smegmatis_MC2_155 DRFSPENSRERD--RWHFIPFAGGARACIGEHFARLETTLALATIIRSVV
Mvan_0858|M.vanbaalenii_PYR-1 DRFSPENVKARD--RWQFLPFLGGGRPCIGEHFARLETTLALATVVRACR
TH_3255|M.thermoresistible__bu DRFAPERFKAMD--RWQFVPFIGGPRSCIGEHFAMLESTLAIATLIRSVR
MMAR_0852|M.marinum_M DRFAPKKSKGRD--RWQYLPFGGGGRSCLGDHFAMLEASLALATIIREAE
MUL_0604|M.ulcerans_Agy99 DRFAPKKSKGRD--RWQYLPFGGGGRSCLGDHFAMLEASLALATIIREAE
MAB_1426|M.abscessus_ATCC_1997 ARFLKENLQPEQ--RKAHMPFGAGKRMCVASGFANLEAIIGIAALAQNYE
MLBr_02088|M.leprae_Br4923 DRPSS-----------RHLAFAVGSHFCLGAALARLEATVTLSAISARFP
* .:.* * : *:. :* * : :: :
Mb1694c|M.bovis_AF2122/97 LQLP-AQRTHRIRAANFAALRPWPGLTVEIRKSAPAQ-------------
Rv1666c|M.tuberculosis_H37Rv LQLP-AQRTHRIRAANFAALRPWPGLTVEIRKSAPAQ-------------
MAV_3106|M.avium_104 LRLP-AQR---LRAANVAALRPTPGLTVEVIDSVPAQ-------------
Mflv_0056|M.gilvum_PYR-GCK VTSS-DNDFPVDVPFTTVAHGPIPAHLTPR--------------------
MSMEG_0968|M.smegmatis_MC2_155 VGSV-DAEFPVDVPFTTVAKGPIRANVAPRGPDHYTL-------------
Mvan_0858|M.vanbaalenii_PYR-1 IESL-DDLFECEVPLTTVAKGPIRARVTPRR-------------------
TH_3255|M.thermoresistible__bu IRSL-DDDFPVEVPFTTVARGPIRAVVSARR-------------------
MMAR_0852|M.marinum_M IHSL-DGDFPVAIPLPIVPAGPIRARVNQRRPSQEVCTTTLSDSSASSSA
MUL_0604|M.ulcerans_Agy99 IHSL-DGDFPVAIPLPIVPAGPIRARVNQRRPSQEVCTTTLSDSSASS--
MAB_1426|M.abscessus_ATCC_1997 LDLLPGQQPRREVTFTGGPEGEILMRLRKRHP------------------
MLBr_02088|M.leprae_Br4923 QVQL-AGELVYKPNVAMRGMSALPVQV-----------------------
Mb1694c|M.bovis_AF2122/97 -
Rv1666c|M.tuberculosis_H37Rv -
MAV_3106|M.avium_104 -
Mflv_0056|M.gilvum_PYR-GCK -
MSMEG_0968|M.smegmatis_MC2_155 -
Mvan_0858|M.vanbaalenii_PYR-1 -
TH_3255|M.thermoresistible__bu -
MMAR_0852|M.marinum_M S
MUL_0604|M.ulcerans_Agy99 -
MAB_1426|M.abscessus_ATCC_1997 -
MLBr_02088|M.leprae_Br4923 -