For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SRITTDQLRHAVLDRGSFVSWDSEPLAVPVADSYARELAAARAATGADESVQTGEGRVFGRRVAVVACEF DFLGGSIGVAAAERITAAVERATAERLPLLASPSSGGTRMQEGTVAFLQMVKIAAAIQLHNQARLPYLVY LRHPTTGGVFASWGSLGHLTVAEPGALIGFLGPRVYELLYGDPFPSGVQTAENLRRHGIIDGVVALDRLR PMLDRALTVLIDAPEPLPAPQTPAPVPDVPTWDSVVASRRPDRPGVRQLLRHGATDRVLLSGTDQGEAAT TLLALARFGGQPTVVLGQQRAVGGGGSTVGPAALREARRGMALAAELCLPLVLVIDAAGPALSAAAEQGG LAGQIAHCLAELVTLDTPTVSILLGQGSGGPALAMLPADRVLAALHGWLAPLPPEGASAIVFRDTAHAAE LAAAQGIRSADLLKSGIVDTIVPEYPDAADEPIEFALRLSNAIAAEVHALRKIPAPERLATRLQRYRRIG LPRD*
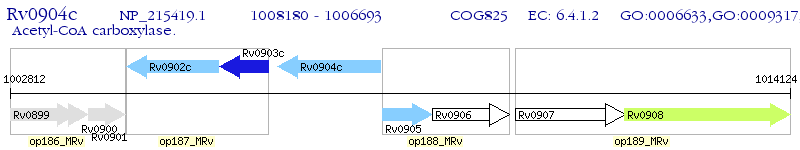
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0904c | accD3 | - | 100% (495) | PUTATIVE ACETYL-COENZYME A CARBOXYLASE CARBOXYL TRANSFERASE (SUBUNIT BETA) ACCD3 (ACCASE BETA CHAIN) |
| M. tuberculosis H37Rv | Rv3280 | accD5 | 1e-09 | 22.99% (461) | propionyl-CoA carboxylase beta chain |
| M. tuberculosis H37Rv | Rv3799c | accD4 | 5e-09 | 22.05% (381) | propionyl-CoA carboxylase beta chain |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0928c | accD3 | 0.0 | 100.00% (495) | putative acetyl-coenzyme A carboxylase carboxyl transferase |
| M. gilvum PYR-GCK | Mflv_1754 | - | 0.0 | 76.83% (492) | carboxyl transferase |
| M. leprae Br4923 | MLBr_00731 | accD5 | 5e-11 | 21.86% (494) | acetyl/propionyl CoA carboxylase beta subunit |
| M. abscessus ATCC 19977 | MAB_4676 | - | 1e-11 | 27.21% (441) | putative carboxyl transferase/pyruvate carboxylase |
| M. marinum M | MMAR_4626 | accD3 | 0.0 | 81.82% (495) | acetyl-coenzyme A carboxylase carboxyl transferase (subunit |
| M. avium 104 | MAV_2190 | - | 5e-10 | 23.28% (378) | propionyl-CoA carboxylase beta chain |
| M. smegmatis MC2 155 | MSMEG_5642 | - | 0.0 | 78.86% (492) | acetyl-coenzyme a carboxylase carboxyl transferase |
| M. thermoresistible (build 8) | TH_0091 | accD3 | 0.0 | 76.16% (495) | PUTATIVE ACETYL-COENZYME A CARBOXYLASE CARBOXYL TRANSFERASE |
| M. ulcerans Agy99 | MUL_0237 | accD3 | 0.0 | 81.41% (495) | acetyl-coenzyme a carboxylase carboxyl transferase (subunit |
| M. vanbaalenii PYR-1 | Mvan_4993 | - | 0.0 | 78.50% (493) | carboxyl transferase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0904c|M.tuberculosis_H37Rv -----------------MSRITTDQLR---------HAVLDRGSFVSWD-
Mb0928c|M.bovis_AF2122/97 -----------------MSRITTDQLR---------HAVLDRGSFVSWD-
MMAR_4626|M.marinum_M -----------------MSRITTDQLR---------DAVLDEGSFISWD-
MUL_0237|M.ulcerans_Agy99 -----------------MSRITTDQLR---------DAVLDEDSFISWD-
Mflv_1754|M.gilvum_PYR-GCK -----------------MSRIGALALR---------DAVLDEGSFLSWD-
Mvan_4993|M.vanbaalenii_PYR-1 -----------------MSRIRALALR---------DALLDEGSFVSWD-
MSMEG_5642|M.smegmatis_MC2_155 -----------------MSRIGALALR---------DAVLDEGSFRSWD-
TH_0091|M.thermoresistible__bu -----------------MSRIGAVQFV---------ETVLDTGSFVSWD-
MLBr_00731|M.leprae_Br4923 MTSVTDHSAHSMERAAEHTINIHTTAGKLAELHKRTEEALHPVGAAAFEK
MAV_2190|M.avium_104 -----------------MTITAPETVG----------ESLDPR-------
MAB_4676|M.abscessus_ATCC_1997 -------MADEQLREDHVELLARRALT---------EDAARPDAVARRHA
Rv0904c|M.tuberculosis_H37Rv ---------SEPLAVPVADSYARELAAARAATGAD-------------ES
Mb0928c|M.bovis_AF2122/97 ---------SEPLAVPVADSYARELAAARAATGAD-------------ES
MMAR_4626|M.marinum_M ---------TPPLEVPISESYARDLADARAATGRD-------------ES
MUL_0237|M.ulcerans_Agy99 ---------TPPLEVPISESYARDLADVRAATGRD-------------ES
Mflv_1754|M.gilvum_PYR-GCK ---------RPPIPVVTSAEYRAELAAAADATGLD-------------ES
Mvan_4993|M.vanbaalenii_PYR-1 ---------GPPLAVPTSAEYRRELAEAEAATGLD-------------ES
MSMEG_5642|M.smegmatis_MC2_155 ---------TAPLDVGADDEYRRELAEASAKTGLD-------------ES
TH_0091|M.thermoresistible__bu ---------DPPLAIPADEDYRRELTDAAAVTGLD-------------EA
MLBr_00731|M.leprae_Br4923 VHAKGKFTARERIYALLDDDSFVELDALARHRSTNF---GLGENRPVGDG
MAV_2190|M.avium_104 -------DPLLRLSNFFDDGSVALLHE--RDRS----------------G
MAB_4676|M.abscessus_ATCC_1997 AGG---RTARENISDLVDAGSFVEYGRFAIAAQRRRRELADLIARTPADG
: .
Rv0904c|M.tuberculosis_H37Rv VQTGEGRVFG-------RRVAVVACEFDFLGGSIGVAAAERITAAVERAT
Mb0928c|M.bovis_AF2122/97 VQTGEGRVFG-------RRVAVVACEFDFLGGSIGVAAAERITAAVERAT
MMAR_4626|M.marinum_M VLTGEGRVLG-------RRVAVVVCEFDFLAGSIGVAAAERITAAVERAT
MUL_0237|M.ulcerans_Agy99 VLTGEGRVLG-------RRVAVVACEFDFLAGSIGVAAAERITAAMERAT
Mflv_1754|M.gilvum_PYR-GCK VVTGEGRVFG-------RRVALIACEFDFLAGSIGVAAAERITAAVQRAT
Mvan_4993|M.vanbaalenii_PYR-1 VLTGEGRVFG-------RRVALIACEFDFLAGSIGVAAAERIIAAVQKAT
MSMEG_5642|M.smegmatis_MC2_155 VLTGEGTVFG-------RRVAVVACEFDFLAGSIGVAAAERITAAVQRAT
TH_0091|M.thermoresistible__bu VLTGEGTIFG-------RRVALVACEFDFLAGSIGVAAAERIVAAVQRAT
MLBr_00731|M.leprae_Br4923 VVTGYGTIDG-------RDVCIFSQDVTVFGGSLGEVYGEKIVKVQELAI
MAV_2190|M.avium_104 VLAAAGTVNG-------VRTIAFCTDGTVMGGAMGIEGCSHIVDAYDTAI
MAB_4676|M.abscessus_ATCC_1997 LVAGTARVNGNLFGADRSACAVLSYDYTVLAGTQGALGHHKKDRLFDLIE
: :. . : * . : .:.*: * : :
Rv0904c|M.tuberculosis_H37Rv AERLPLLASPSSGGTRMQEGTVAFLQMVKI-AAAIQLHNQARLPYLVYLR
Mb0928c|M.bovis_AF2122/97 AERLPLLASPSSGGTRMQEGTVAFLQMVKI-AAAIQLHNQARLPYLVYLR
MMAR_4626|M.marinum_M RQRLPLLASPSSGGTRMQEGTVAFLQMVKI-AAAVRLHKQAHLPYLVYLR
MUL_0237|M.ulcerans_Agy99 SQRLPLLASPSSGGTRMQEGTVAFLQMVKI-AAAVRLHKQAHLPYLVYLR
Mflv_1754|M.gilvum_PYR-GCK AEGLPLLASPSSGGTRMQEGTVAFLQMVKI-AAAVELHKKGNLPYLVYLR
Mvan_4993|M.vanbaalenii_PYR-1 AERLPLLASPSSGGTRMQEGTVAFLQMVKI-AAAVELHKKAHLPYLVYLR
MSMEG_5642|M.smegmatis_MC2_155 AERLPLLASPSSGGTRMQEGTVAFLQMVKI-AAAVELHKQAHLPYLVYLR
TH_0091|M.thermoresistible__bu AERLPLVASPSSGGTRMQEGTVAFLQMVKI-AAAVQLHKLAHLSYLVYLR
MLBr_00731|M.leprae_Br4923 KTGRPLIGINDGAGARIQEG-VVSLGLYSR-IFRNNILASGVIPQISLIM
MAV_2190|M.avium_104 EEQSPIVGIWHSGGARLAEG-VKALHAVGL-VFEAMIRASGYIPQISVVV
MAB_4676|M.abscessus_ATCC_1997 RMKLPTVFFAEGGGGRPGDTDYPVVSMLDVRAFKLWAALSGVVPRISVVN
* : ..* * : : . :. : :
Rv0904c|M.tuberculosis_H37Rv HPTTGGVFASWGSLGHLTVAEPGALIGFLGPRVYELLYGDPFPSGVQTAE
Mb0928c|M.bovis_AF2122/97 HPTTGGVFASWGSLGHLTVAEPGALIGFLGPRVYELLYGDPFPSGVQTAE
MMAR_4626|M.marinum_M HPTTGGVFASWGSLGHVTVAEPGALIGFLGPRVYELLYDEPFPAGIQTAE
MUL_0237|M.ulcerans_Agy99 HPTTGGVFASWGSLGHVTVAEPGALIGFLGPRVYELLYDEPFPAGVQTAE
Mflv_1754|M.gilvum_PYR-GCK HPTTGGVFASWGSLGHVTAAEPDALIGFLGPRVYEHLYGERFPSGVQTSE
Mvan_4993|M.vanbaalenii_PYR-1 HPTTGGVFASWGSLGHVTAAEPGALIGFLGPRVYEHLYGEPFPAGIQTAE
MSMEG_5642|M.smegmatis_MC2_155 HPTTGGVFASWGSLGHVTAAEPGALIGFLGPRVYEHLYGEPFPSGVQTAE
TH_0091|M.thermoresistible__bu HPTTGGVFASWGSLGHITIAEPNALIGFLGPRVYQHLYGEPFPPGVQTAE
MLBr_00731|M.leprae_Br4923 GAAAGGHVYSPALTDFVVMVDQTSQMFITGPDVIKTVTGEDVTMEELGGA
MAV_2190|M.avium_104 GFAAGGAAYGPALTDVVVMAPE-SRVFVTGPDVVRSVTGEDVDMASLGGP
MAB_4676|M.abscessus_ATCC_1997 GRCFAGNAVIAGCS-DLIVATKDTSIGMGGPAMIAGGGLGEVHPDEVGPI
.* . : . : : . ** : .
Rv0904c|M.tuberculosis_H37Rv NLRRHGIIDGVVALDRLRPMLDRALTVLIDAPEPLPAPQTPA--------
Mb0928c|M.bovis_AF2122/97 NLRRHGIIDGVVALDRLRPMLDRALTVLIDAPEPLPAPQTPA--------
MMAR_4626|M.marinum_M NLQRHGVIDGVVTLDELRRTLDRALTVLTDAPAPAPAAPAPT--------
MUL_0237|M.ulcerans_Agy99 NLQRHGVIDGVVTLDELRRTLDRALTVLTDAPAPAPAAPAPP--------
Mflv_1754|M.gilvum_PYR-GCK NLQRHGVIDAVVPLEVLRATVDRTLKVVADAPGPPPAAPEPE--------
Mvan_4993|M.vanbaalenii_PYR-1 NLHRHGVVDAVIPPGILRATLDRALRVVSDAPDVPPAAPDAE--------
MSMEG_5642|M.smegmatis_MC2_155 NLHAHGVIDGVVPLALLRETLDRTLRVVADPPGPAPAVIETE--------
TH_0091|M.thermoresistible__bu NLHRHGVIDSVVPLPRLRSELDRALRVLVDEPGTPPAPPDVDT-------
MLBr_00731|M.leprae_Br4923 HTHMAKSGTAHYVASGEQDAFDWVRDVLSYLPSNNFTDAPRYSKPVPHGS
MAV_2190|M.avium_104 ETHHKKSGVCHIVADDELDAYERGRRLVGLFCQQGHFDR-----------
MAB_4676|M.abscessus_ATCC_1997 AVQSTN-GVVDVVVDDEEQAVTVAKRLISYFQG----------------A
: ::
Rv0904c|M.tuberculosis_H37Rv PVPDVPTWDSVVASRRPDRP----GVRQLLRHGATDRVLLSGTDQGEAAT
Mb0928c|M.bovis_AF2122/97 PVPDVPTWDSVVASRRPDRP----GVRQLLRHGATDRVLLSGTDQGEAAT
MMAR_4626|M.marinum_M AIPDVPAWTSVLASRRPDRP----GVEHLLRYGATDRVLLSGTGYGDAAT
MUL_0237|M.ulcerans_Agy99 AIPDVAAWTSVLASRRPDRP----GVEHLLRYGATDRVLLSGTGYGDAAT
Mflv_1754|M.gilvum_PYR-GCK VLPDIPAWDSVEISRRPDRP----GVSFLLRHGATERVLLSGTGQGESAT
Mvan_4993|M.vanbaalenii_PYR-1 PLPDVPAWESVEISRRPDRP----GVSALLRYGTTDRVLLSGTGQGEAAT
MSMEG_5642|M.smegmatis_MC2_155 DLPDVPAWDSVIASRRPDRP----GVGFLLRHGATDQVLLSGTERGEAAT
TH_0091|M.thermoresistible__bu SQPDVPAWDSVIASRRPDRP----GVGYLLRHGATDTVLLSGSGGGEAAT
MLBr_00731|M.leprae_Br4923 IEDNLTAKDLELDTLIPDSPNQPYDMHEVVTRLLDEEE-FLEVQAGYATN
MAV_2190|M.avium_104 --TKAEAGDTDIHALLPESARRAYDVRPIVTAILDDETPFDEFQANWAPS
MAB_4676|M.abscessus_ATCC_1997 VAPGDGADQTALRTMIPERARRAYPIRPIIET-LADTGSVTFLREKFAPE
: : *: . : :: : . :.
Rv0904c|M.tuberculosis_H37Rv TLLALARFGGQPTVVLGQQRAVGGGGSTVGPAALREARRGMALAAELCLP
Mb0928c|M.bovis_AF2122/97 TLLALARFGGQPTVVLGQQRAVGGGGSTVGPAALREARRGMALAAELCLP
MMAR_4626|M.marinum_M TLLALARMAGQPMVVLGQQ-GIGGG--IIGPASLREARRGMALAAELRLP
MUL_0237|M.ulcerans_Agy99 TLLALARMAGQPMVVLGQQ-GIGGG--IIGPASLREARRGMALAAELRLP
Mflv_1754|M.gilvum_PYR-GCK MMLALARFGNQPAVVLGQQ-RVVGG--LVGPAALREARRGMALAASLKLP
Mvan_4993|M.vanbaalenii_PYR-1 TLLALARFGGQPAVVLGQQ-RVVGG--LVGPAALREARRGMALAAGLKLP
MSMEG_5642|M.smegmatis_MC2_155 TLLALARFGGQPAVVLGQR-RIVGG--LVGPGALREARRGMALAAGLQLP
TH_0091|M.thermoresistible__bu TLLALARFGGQPAVVLGQQ-RVIGA--MVGPAALREARRGMALAADLQLP
MLBr_00731|M.leprae_Br4923 IVVGLGRIDDRPVGIVANQPIQFAG--CLDINASEKAARFVRVCDCFNIP
MAV_2190|M.avium_104 MVIGLGRLSGRTVGVLANNPLRLGG--CLNSESAEKAARFVRLCDAFGIP
MAB_4676|M.abscessus_ATCC_1997 MATALARIEGRPIGVLANNSMVMAG--AITAAASDKAARFLQLCDAFGLP
.*.*: .:. ::.:. .. : : :* * : :. : :*
Rv0904c|M.tuberculosis_H37Rv LVLVIDAAGPALSAAAEQGGLAGQIAHCLAELVTLDTPTVSILLGQGSGG
Mb0928c|M.bovis_AF2122/97 LVLVIDAAGPALSAAAEQGGLAGQIAHCLAELVTLDTPTVSILLGQGSGG
MMAR_4626|M.marinum_M LVLVIDTAGPALSADAEQGGLAGQIAQCLAELVTLGTPTVSVLLGQGSGG
MUL_0237|M.ulcerans_Agy99 LVLVIDTAGPALSADAEQGGLAGQIAQCLAELVTLGTPTVSVLLGQGSGG
Mflv_1754|M.gilvum_PYR-GCK LVLVIDTAGPALSTEAEEGGLAGEIARCLAELVTLDTPTVSVLLGQGSGG
Mvan_4993|M.vanbaalenii_PYR-1 LVLVIDTAGPALSTEAEEGGLAGEIARCLAELVTLDTPTVSVLLGQGSGG
MSMEG_5642|M.smegmatis_MC2_155 LVLVIDTAGPALSTEAEQGGLAGEIARCLAELVTLDTPTVSVLMGQGSGG
TH_0091|M.thermoresistible__bu LVLVIDTAGPALTAEAEQGGLASEIAHCLAELVTLDTATLSVLLGQGSGG
MLBr_00731|M.leprae_Br4923 IVMLVDVPGFLPGTEQEYDGIIRRGAKLLFAYGEATVPKITVITRKAYGG
MAV_2190|M.avium_104 LVVIVDVPGYLPGVDQEWGGVVRRGAKLLHAFGEATVPRVTLVTRKTYGG
MAB_4676|M.abscessus_ATCC_1997 VLSLVDCPGYMVGPAAEAEALVRRASRMLVAGAALRVPLVSVILRRGYGL
:: ::* .* * .: . :: * .. :::: : *
Rv0904c|M.tuberculosis_H37Rv PALAMLP-----ADRVLAALHGWLAPLPPEGASAIVFRDTAHAAELAAAQ
Mb0928c|M.bovis_AF2122/97 PALAMLP-----ADRVLAALHGWLAPLPPEGASAIVFRDTAHAAELAAAQ
MMAR_4626|M.marinum_M PALAMVP-----ADRVLAAQHGWLAPLPPEGASAIVFRDTTHAAELAAAQ
MUL_0237|M.ulcerans_Agy99 PALAMVP-----ADRVLAAQHGWLAPLPPEGASAIVFRDTTHAAELAAAQ
Mflv_1754|M.gilvum_PYR-GCK PALAMVP-----ADRVLAALHGWLAPLPPEGASAIVFRDVDHAPELAAAQ
Mvan_4993|M.vanbaalenii_PYR-1 PALAMVP-----ADRVLAALHGWLAPLPPEGASAIVFRDVDHAPELAAAQ
MSMEG_5642|M.smegmatis_MC2_155 PALAMVP-----ADRVLAASNGWLAPLPPEGASAIVYRDTEHAPELAAAQ
TH_0091|M.thermoresistible__bu PALAMVP-----ADRVLAARHGWLAPLPPEGVSAIVHRDLDHAPELAARQ
MLBr_00731|M.leprae_Br4923 AYCVMGSK-NMGCDVNLAWPTAQIAVMGASGAVGFVYRKELAQAAKNGAN
MAV_2190|M.avium_104 AYIAMNSR-SLNATKVFAWPDAEVAVMGAKAAVGILHKKKLAAAPEHER-
MAB_4676|M.abscessus_ATCC_1997 GAQAMTGGSLHEPLLTVAWPNAHLGPMGLEGAVRLGLRKELEAIADESAR
.* .* . :. : ... : :.
Rv0904c|M.tuberculosis_H37Rv GIRSADLLKSGIVDTIVPEYPDAADEPIEFALRLSNAIAAEVHALRKIPA
Mb0928c|M.bovis_AF2122/97 GIRSADLLKSGIVDTIVPEYPDAADEPIEFALRLSNAIAAEVHALRKIPA
MMAR_4626|M.marinum_M GIRSADLLAAGIVDVIVPELPDAAAEPVEFAQRLSIAIATEVAALREIPD
MUL_0237|M.ulcerans_Agy99 GIRSADLLAAGIVDVIVPELPDAAAEPVEFAQRLSIAIATEVAALREIPD
Mflv_1754|M.gilvum_PYR-GCK GIRSVDLLRDGIVDAVVPEHPDAADEPRAFVERLSATIAVELHRLRFAPE
Mvan_4993|M.vanbaalenii_PYR-1 GIRSADLLRNGIVDAIVPEHPDAADEPQAFTQRLSATIAVELHRLRTVPD
MSMEG_5642|M.smegmatis_MC2_155 GVRAVDLLRHGIVDVVVAEKPDAADEPRAFTARLSTAIAAELHNLRAVPD
TH_0091|M.thermoresistible__bu GIRSVDLLRHGIVDAIVDERPDAADEPMEFTARLADSIAVELHRLREMPA
MLBr_00731|M.leprae_Br4923 VDELRLQLQQEYEDTLVNPYIAAERGYVDAVIPPSHTRGYIATALHLLER
MAV_2190|M.avium_104 -EALHDQLAAEHEKIAGGVDSAIEIGVVDEKIDPAHTRSKLTEALAQAPA
MAB_4676|M.abscessus_ATCC_1997 EEAVRQATAAAQENAKAIN--AAQIFEIDDVIDPAETRSLVASTFAAAMR
. : : . :
Rv0904c|M.tuberculosis_H37Rv PERLATRLQRYRRIGLPRD-
Mb0928c|M.bovis_AF2122/97 PERLATRLQRYRRIGLPRD-
MMAR_4626|M.marinum_M AQRMTARLARYRSIGLPRDR
MUL_0237|M.ulcerans_Agy99 AQRMTARLARYRSIGLPRDR
Mflv_1754|M.gilvum_PYR-GCK AERSASRLERYRRIGLA---
Mvan_4993|M.vanbaalenii_PYR-1 EQRLAARLERYRRIGLP---
MSMEG_5642|M.smegmatis_MC2_155 DERLQTRLQRYRRIGL----
TH_0091|M.thermoresistible__bu ESRLETRLARYRRIGLPR--
MLBr_00731|M.leprae_Br4923 KIAHLP-PKKHGNIPL----
MAV_2190|M.avium_104 RRG------RHKNIPL----
MAB_4676|M.abscessus_ATCC_1997 EPLPLRRSVVDTW-------