For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTAPTGTSATTTRPWTPRIATQLSVLACAAFIYVTAEILPVGALSAIARNLRVSVVLVGTLLSWYALVAA VTTVPLVRWTAHWPRRRALVVSLVCLTVSQLVSALAPNFAVLAAGRVLCAVTHGLLWAVIAPIATRLVPP SHAGRATTSIYIGTSLALVVGSPLTAAMSLMWGWRLAAVCVTGAAAAVALAARLALPEMVLRADQLEHVG RRARHHRNPRLVKVSVLTMIAVTGHFVSYTYIVVIIRDVVGVRGPNLAWLLAAYGVAGLVSVPLVARPLD RWPKGAVIVGMTGLTAAFTLLTALAFGERHTAATALLGTGAIVLWGALATAVSPMLQSAAMRSGGDDPDG ASGLYVTAFQIGIMAGALLGGLLYERSLAMMLTASAGLMGVALFGMTVSQHLFENPTLSPGDG
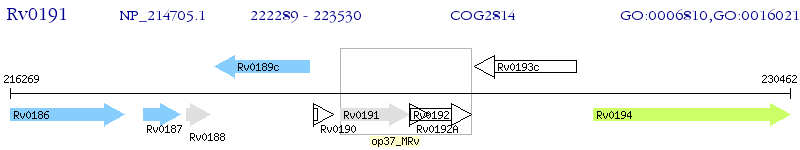
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0191 | - | - | 100% (413) | PROBABLE CONSERVED INTEGRAL MEMBRANE PROTEIN |
| M. tuberculosis H37Rv | Rv0783c | emrB | 3e-07 | 23.36% (214) | multidrug resistance integral membrane efflux protein EmrB |
| M. tuberculosis H37Rv | Rv1634 | - | 6e-07 | 27.55% (196) | drug efflux membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0197 | - | 0.0 | 100.00% (413) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_1840 | - | 2e-33 | 30.30% (396) | major facilitator transporter |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1429 | - | 6e-30 | 29.34% (409) | major facilitator superfamily permease |
| M. marinum M | MMAR_0434 | - | 0.0 | 80.58% (412) | integral membrane protein |
| M. avium 104 | MAV_4987 | - | 1e-176 | 76.46% (412) | sugar transporter family protein |
| M. smegmatis MC2 155 | MSMEG_0232 | - | 1e-126 | 59.95% (397) | sugar transporter family protein |
| M. thermoresistible (build 8) | TH_3988 | - | 1e-29 | 30.11% (269) | PUTATIVE putative MFS permease |
| M. ulcerans Agy99 | MUL_1084 | - | 0.0 | 80.58% (412) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_4890 | - | 6e-32 | 29.82% (399) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0191|M.tuberculosis_H37Rv -------MTAPTGTSATTTRPWTPRIATQLSVLACAAFIYVTAEILPVGA
Mb0197|M.bovis_AF2122/97 -------MTAPTGTSATTTRPWTPRIATQLSVLACAAFIYVTAEILPVGA
MMAR_0434|M.marinum_M -------MTSEARTATTAARPWTPRVAAQLSVLAAAAFIYVTAEILPVGA
MUL_1084|M.ulcerans_Agy99 -------MTSEARTATTAARPWTPRVAAQLSVLAAAAFIYVTAEILPVGA
MAV_4987|M.avium_104 -------MTAET--ANAAARTWTPRIAAQLAILAAAAFTYVTAEILPVGA
MSMEG_0232|M.smegmatis_MC2_155 -------MTVTP----PTVSPWTVRVAAQLTVLAAAAFIYVTAEILPVGA
Mflv_1840|M.gilvum_PYR-GCK ---MSAAVDQQSTPGASVAAPDPVVRRLAVLALALGGFGIGTTEFVAMGL
Mvan_4890|M.vanbaalenii_PYR-1 ---MTDTLHRPSGP-VSVAAPDPVVRRLAVVALALGGFGIGTTEFVAMGL
MAB_1429|M.abscessus_ATCC_1997 ---MTAVVDSRVQP---AAPVSPRVRRGAILALALGGFGIGTTEFVAMGL
TH_3988|M.thermoresistible__bu MSESKASQDASAPAADPVDATARRTPWIAILAAALGIFVMITIEELPIGV
. : * . * * * :.:*
Rv0191|M.tuberculosis_H37Rv LSAIARNLRVSVVLVGTLLSWYALVAAVTTVPLVRWTAHWPRRRALVVSL
Mb0197|M.bovis_AF2122/97 LSAIARNLRVSVVLVGTLLSWYALVAAVTTVPLVRWTAHWPRRRALVVSL
MMAR_0434|M.marinum_M LSAIARNLHISVVLVGTLLSWYALVAALTTVPLVRWTAHWPRRHALMVSL
MUL_1084|M.ulcerans_Agy99 LSAIARNLHISVVLVGTLLSWYALVAALTTVPLVRWTAHWPRRHALMVSL
MAV_4987|M.avium_104 LPAIARNLQVSLVLVGTLLSWYALVAALTTIPLVRWTAHLPRRRVLVASL
MSMEG_0232|M.smegmatis_MC2_155 LPVIAADLNVSEGLVGTLMAGYALIAALTTVALVRLTARWPRRRALLVTM
Mflv_1840|M.gilvum_PYR-GCK LPDIARSFDVSEPVAGHVISAYALGVVVGAPLIAAITARMARRTLLLALM
Mvan_4890|M.vanbaalenii_PYR-1 LPDIATGMGVSEPTAGHVISAYALGVVVGAPVIAAVTARMARRKLLLALM
MAB_1429|M.abscessus_ATCC_1997 LPNIAHGLHVTEPQAGHVISAYALGVVVGAPVIAALTARASRRTLLVALM
TH_3988|M.thermoresistible__bu LTLIGDDLGLSSGVVGLTVTIPGVLAGAVAVLTPTLIGRLDRRLALVSAL
*. *. .: :: .* :: .: . : .: ** *: :
Rv0191|M.tuberculosis_H37Rv VCLTVSQLVSALAPNFAVLAAGRVLCAVTHGLLWAVIAPIATRLVPPSHA
Mb0197|M.bovis_AF2122/97 VCLTVSQLVSALAPNFAVLAAGRVLCAVTHGLLWAVIAPIATRLVPPSHA
MMAR_0434|M.marinum_M VCLTISQLISALAPNFAVLAAGRALCAITHGLLWSVIAPIATRLVPPSHA
MUL_1084|M.ulcerans_Agy99 VCLTISQLISALAPNFAVLAAGRALCAITHGLLWSVIAPIATRLVPPSHA
MAV_4987|M.avium_104 TCLTASQLISALAPNFAVLAAGRVLCAITHGLLWSVIAPIATRLVPPSHA
MSMEG_0232|M.smegmatis_MC2_155 VCLTVSQVISAMAPNFAVLAGGRVLCALAHGLMWSVIAPIGVRLVPPSHA
Mflv_1840|M.gilvum_PYR-GCK AVFTLGNLASMLASSYETLVGARFLAGLPHGAYFGVAALVAAHLMGPQHR
Mvan_4890|M.vanbaalenii_PYR-1 ALFTIGNLASMLAPTYETLIAARFLAGLPHGAYFGVAALVAAHLMGPQNR
MAB_1429|M.abscessus_ATCC_1997 VAFTIGNAATVFAPGYAALIASRFVAGLPHGAYFGVAALVAAHLAGPGER
TH_3988|M.thermoresistible__bu LLVFVSAATSAIAPNLPVLLASRLFAGMAIGVFWALLAVVVARVAHPDDL
. . : :*. .* ..* ...:. * :.: * : .:: * .
Rv0191|M.tuberculosis_H37Rv GRATTSIYIGTSLALVVGSPLTAAMSLMWGWRLAAVCVTGAAAAVALAAR
Mb0197|M.bovis_AF2122/97 GRATTSIYIGTSLALVVGSPLTAAMSLMWGWRLAAVCVTGAAAAVALAAR
MMAR_0434|M.marinum_M GRATTSIYIGTSLALVIGSPLTAALSLMWGWRLAAVCVTVAAAVVTVAAR
MUL_1084|M.ulcerans_Agy99 GRATTSIYIGTSLALVIGSPLTAALSLMWGWRLAAVCVTVAAAVVTVAAR
MAV_4987|M.avium_104 GRATMSIYVGTSLALVVGSPLTAALSLMWGWRLAVVCVTVAAAVVTVAAR
MSMEG_0232|M.smegmatis_MC2_155 GRATAAVYVGTGLALVVGNPLTAALSELWGWRQAVVAVAIAAAAVTLAAR
Mflv_1840|M.gilvum_PYR-GCK AKAVAHVLTGLTVATVLGVPVASWLGQSLGWR-AAFGLVVAVGVLTLTAL
Mvan_4890|M.vanbaalenii_PYR-1 AKAVAHVLTGLTVATVLGVPIASWLGQSLGWR-AAFGLVVGVGLVTLTAL
MAB_1429|M.abscessus_ATCC_1997 AKAVGQTMLGLSVANVVGVPVATWLGNTVGWR-SAFAVVAVIGAITIAAL
TH_3988|M.thermoresistible__bu PKALTLAFGGVSVAVVLGVPFAAWIGAMVGWRWAFVLVGALGLMVAVALT
:* * :* *:* *.:: :. *** : . : ::::
Rv0191|M.tuberculosis_H37Rv LALPEMVLRADQLEHVGRRARHHRNPRLVKVSVLTMIAVTGHFVSYTYIV
Mb0197|M.bovis_AF2122/97 LALPEMVLRADQLEHVGRRARHHRNPRLVKVSVLTMIAVTGHFVSYTYIV
MMAR_0434|M.marinum_M LLLPEMVLSADQLQYVGPRSRHHRNRALIIVSLITMVGVTGHFVSYTYIV
MUL_1084|M.ulcerans_Agy99 LLLPEMVLSADQLQHVGPRSRHHRNRALIIVSLITMVGVTGHFVSYTYIV
MAV_4987|M.avium_104 LMLPEMVLTEHQLAHVGPRSRHHRNGRLITVSLLAMVAVTGHFVSYTFIV
MSMEG_0232|M.smegmatis_MC2_155 VLLPLMPAERARAGSAG-RPVHGRG--LLTLSLLTLIGVTGHFVAYTFIV
Mflv_1840|M.gilvum_PYR-GCK ALWLPVQLRLMRSTSPLTELGALRRPQVWLALAVGMIGFGGMFAVYTYIA
Mvan_4890|M.vanbaalenii_PYR-1 WCWLPFQLKFMRATSPLTELGALRRPQVWLALLVGMIGFGGMFAVYTYIT
MAB_1429|M.abscessus_ATCC_1997 LRYLP-GLTDMRITNPLTELGALRRVQVWFTLLIGIVGFGGMFAFYTYIS
TH_3988|M.thermoresistible__bu VLLPPVPVAER--TTLGQLGAAWSIPAARTGVLVTLVMVTAHFTAYTYAS
: :: . . *. **:
Rv0191|M.tuberculosis_H37Rv VIIRDVVGVRGPNLAWLLAAYGVAGLVSVPLVARPLDRWPKGAVIVGMTG
Mb0197|M.bovis_AF2122/97 VIIRDVVGVRGPNLAWLLAAYGVAGLVSVPLVARPLDRWPKGAVIVGMTG
MMAR_0434|M.marinum_M VIIRQVVGVRGPSLAWLLAAYGVAGVVAVALVARPLDRRPKGTIIFCVAG
MUL_1084|M.ulcerans_Agy99 VIIRQVVGVRGPSLAWLLAAYGVAGVVAVALVARPLDRRPKGTIIFCVAG
MAV_4987|M.avium_104 EIIRNVVGVRGPTLAWVLAAYGLAGLLSVPLVARPLDHRPKSAVILCMTG
MSMEG_0232|M.smegmatis_MC2_155 AIIRDVVGIDGPHLAWLLAGYGVAGLAAMAAMARPLDLWPKASVTAALVV
Mflv_1840|M.gilvum_PYR-GCK TTMTDVAGMPRALVPVALMVFGLGMFFGNLVGGRLADRSVVRALYLSMGA
Mvan_4890|M.vanbaalenii_PYR-1 TTMTDVAGMPRGLVPLALMMFGLGMVLGNLVGGRLADGSVVRALYLSLGA
MAB_1429|M.abscessus_ATCC_1997 TTLTSVSGLPRALVPLALSLYGIGMVTGNVLGGWMADRSVVRAIAVGLVL
TH_3988|M.thermoresistible__bu PVLQDRIHITESGVGPMLFVFGIAGFLGNFAAGPLLRRSMRATYVVLPIG
: . : : * :*:. . . . :
Rv0191|M.tuberculosis_H37Rv LTAAFTLLTALAFGERHTAAT--ALLG-TGAIVLWGALATAVSPMLQSAA
Mb0197|M.bovis_AF2122/97 LTAAFTLLTALAFGERHTAAT--ALLG-TGAIVLWGALATAVSPMLQSAA
MMAR_0434|M.marinum_M LTFAFVLLTALAFGGHLAPMT--ALVVGTGAIVLWGAAATAVSPMLQSAA
MUL_1084|M.ulcerans_Agy99 LTFAFVLLTALAFGGHLAPMT--ALVVGTGAIVLWGAAVTAVSPMLQSAA
MAV_4987|M.avium_104 LTAAFAVLTALAFGGPTGATT--ALIG-TAAIVLWGAMATAVSPMMQSAA
MSMEG_0232|M.smegmatis_MC2_155 LAASLVTLAVLSAQPRAGVTTGAVMVIGTVAIVLWGASSTALPPMLQTSA
Mflv_1840|M.gilvum_PYR-GCK LCAMLTLFVAASHNPWTALLA----------LFGIGLTGSAVGPALQTRL
Mvan_4890|M.vanbaalenii_PYR-1 LCGALALFVVASHNPWTALLV----------LFLIGLTGSAVGPALQTRL
MAB_1429|M.abscessus_ATCC_1997 VAAMLLIFAAVSRNPWAALAL----------VFAVGVSGTSLVPALQTRL
TH_3988|M.thermoresistible__bu VMVGVXXXGPNSRWRPDGTRM-----------------------------
: . :
Rv0191|M.tuberculosis_H37Rv MRSGGDDPDGASGLYVTAFQIGIMAGALLGGLLYERS-LAMMLTASAGLM
Mb0197|M.bovis_AF2122/97 MRSGGDDPDGASGLYVTAFQIGIMAGALLGGLLYERS-LAMMLTASAGLM
MMAR_0434|M.marinum_M MRSGADDPDGASGLYVTAFQVGIMAGSLIGGLLYERS-VAMMLTASAGLM
MUL_1084|M.ulcerans_Agy99 MRSGADDPDGASGLYVTAFQVGIMAGSLIGGLLYERS-VAMMLTASAGLM
MAV_4987|M.avium_104 MRNGADDPDGASGLYVTAFQVGIMAGSLLGGLLYERS-VILMLSASGVLM
MSMEG_0232|M.smegmatis_MC2_155 MRTSPEDPDGASGRYVAAFQVGIMTGALVGAGVYEAVGLTAMIGTAAVLI
Mflv_1840|M.gilvum_PYR-GCK MDVAHDAQTLAAALNHSALNIGNATGAWVGGLVIAAGYGYTAPAGAGAVL
Mvan_4890|M.vanbaalenii_PYR-1 MDVAHDAQTLAAALNHSALNIGNATGAWVGGLVIAAGLGYTAPAAAGAVL
MAB_1429|M.abscessus_ATCC_1997 MDVADDAQTLAAALNHSALNIANAGGAWLGGLVITAGYGYRAPSLVGAAL
TH_3988|M.thermoresistible__bu --------------------------------------------------
Rv0191|M.tuberculosis_H37Rv GVALFGMTVSQHLFENPTLSPGDG----
Mb0197|M.bovis_AF2122/97 GVALFGMTVSQHLFENPTLSPGDG----
MMAR_0434|M.marinum_M GVALVGMTSARRVFEAPVAVAGATSHNQ
MUL_1084|M.ulcerans_Agy99 GVALVGMTSARRVFEAPVAVAGATSHNQ
MAV_4987|M.avium_104 AVALVGIAANRRMLDVAPTSSRDS----
MSMEG_0232|M.smegmatis_MC2_155 TAAMCGALAVPDVFNRSRDLSTR-----
Mflv_1840|M.gilvum_PYR-GCK ALAGLVVLTVSVVLQKR-----------
Mvan_4890|M.vanbaalenii_PYR-1 ALAGLAVLTVSVLLQKRG----------
MAB_1429|M.abscessus_ATCC_1997 AIAGLVVLGLSVLYARSRAVALPSRAG-
TH_3988|M.thermoresistible__bu ----------------------------