For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTVTPPTVSPWTVRVAAQLTVLAAAAFIYVTAEILPVGALPVIAADLNVSEGLVGTLMAGYALIAALTTV ALVRLTARWPRRRALLVTMVCLTVSQVISAMAPNFAVLAGGRVLCALAHGLMWSVIAPIGVRLVPPSHAG RATAAVYVGTGLALVVGNPLTAALSELWGWRQAVVAVAIAAAAVTLAARVLLPLMPAERARAGSAGRPVH GRGLLTLSLLTLIGVTGHFVAYTFIVAIIRDVVGIDGPHLAWLLAGYGVAGLAAMAAMARPLDLWPKASV TAALVVLAASLVTLAVLSAQPRAGVTTGAVMVIGTVAIVLWGASSTALPPMLQTSAMRTSPEDPDGASGR YVAAFQVGIMTGALVGAGVYEAVGLTAMIGTAAVLITAAMCGALAVPDVFNRSRDLSTR
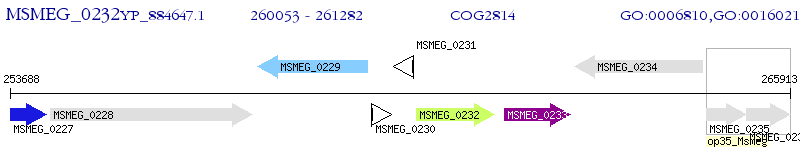
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0232 | - | - | 100% (409) | sugar transporter family protein |
| M. smegmatis MC2 155 | MSMEG_3314 | - | 1e-40 | 33.16% (386) | transport protein |
| M. smegmatis MC2 155 | MSMEG_2028 | - | 4e-37 | 30.18% (381) | chloramphenicol resistance protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0197 | - | 1e-126 | 59.95% (397) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_1840 | - | 2e-32 | 30.13% (385) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv0191 | - | 1e-126 | 59.95% (397) | integral membrane protein |
| M. leprae Br4923 | MLBr_01562 | - | 6e-07 | 23.11% (264) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | MAB_1429 | - | 8e-30 | 27.97% (404) | major facilitator superfamily permease |
| M. marinum M | MMAR_0434 | - | 1e-127 | 59.64% (394) | integral membrane protein |
| M. avium 104 | MAV_4987 | - | 1e-123 | 59.79% (388) | sugar transporter family protein |
| M. thermoresistible (build 8) | TH_3988 | - | 3e-34 | 33.57% (280) | PUTATIVE putative MFS permease |
| M. ulcerans Agy99 | MUL_1084 | - | 1e-127 | 59.64% (394) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_4890 | - | 4e-31 | 28.99% (376) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1840|M.gilvum_PYR-GCK --------------------------------MSAAVDQQSTPGASVAAP
Mvan_4890|M.vanbaalenii_PYR-1 --------------------------------MTDTLHRPSGP-VSVAAP
MAB_1429|M.abscessus_ATCC_1997 --------------------------------MTAVVDSRVQP---AAPV
Mb0197|M.bovis_AF2122/97 ------------------------------------MTAPTGTSATTTRP
Rv0191|M.tuberculosis_H37Rv ------------------------------------MTAPTGTSATTTRP
MMAR_0434|M.marinum_M ------------------------------------MTSEARTATTAARP
MUL_1084|M.ulcerans_Agy99 ------------------------------------MTSEARTATTAARP
MAV_4987|M.avium_104 ------------------------------------MTAET--ANAAART
MSMEG_0232|M.smegmatis_MC2_155 ------------------------------------MTVTP----PTVSP
TH_3988|M.thermoresistible__bu -----------------------------MSESKASQDASAPAADPVDAT
MLBr_01562|M.leprae_Br4923 MVTLYDHERAVHNWTAARSDRPAPSRLTQQVEPASERTSKYPTLLPSGRF
Mflv_1840|M.gilvum_PYR-GCK DPVVRRLAVLALALGGFGIGTTEFVAMGLLPDIARSFDVSEPVAGHVISA
Mvan_4890|M.vanbaalenii_PYR-1 DPVVRRLAVVALALGGFGIGTTEFVAMGLLPDIATGMGVSEPTAGHVISA
MAB_1429|M.abscessus_ATCC_1997 SPRVRRGAILALALGGFGIGTTEFVAMGLLPNIAHGLHVTEPQAGHVISA
Mb0197|M.bovis_AF2122/97 WTPRIATQLSVLACAAFIYVTAEILPVGALSAIARNLRVSVVLVGTLLSW
Rv0191|M.tuberculosis_H37Rv WTPRIATQLSVLACAAFIYVTAEILPVGALSAIARNLRVSVVLVGTLLSW
MMAR_0434|M.marinum_M WTPRVAAQLSVLAAAAFIYVTAEILPVGALSAIARNLHISVVLVGTLLSW
MUL_1084|M.ulcerans_Agy99 WTPRVAAQLSVLAAAAFIYVTAEILPVGALSAIARNLHISVVLVGTLLSW
MAV_4987|M.avium_104 WTPRIAAQLAILAAAAFTYVTAEILPVGALPAIARNLQVSLVLVGTLLSW
MSMEG_0232|M.smegmatis_MC2_155 WTVRVAAQLTVLAAAAFIYVTAEILPVGALPVIAADLNVSEGLVGTLMAG
TH_3988|M.thermoresistible__bu ARRTPWIAILAAALGIFVMITIEELPIGVLTLIGDDLGLSSGVVGLTVTI
MLBr_01562|M.leprae_Br4923 PSGRFIAAVIVIGGMQLLATMDSTVAIVALPKIQNELSLSDAGRSWGITA
: . : . :.: *. * : :: . ::
Mflv_1840|M.gilvum_PYR-GCK YALGVVVGAPLIAAITARMARRTLLLALMAVFTLGNLASMLASSYETLVG
Mvan_4890|M.vanbaalenii_PYR-1 YALGVVVGAPVIAAVTARMARRKLLLALMALFTIGNLASMLAPTYETLIA
MAB_1429|M.abscessus_ATCC_1997 YALGVVVGAPVIAALTARASRRTLLVALMVAFTIGNAATVFAPGYAALIA
Mb0197|M.bovis_AF2122/97 YALVAAVTTVPLVRWTAHWPRRRALVVSLVCLTVSQLVSALAPNFAVLAA
Rv0191|M.tuberculosis_H37Rv YALVAAVTTVPLVRWTAHWPRRRALVVSLVCLTVSQLVSALAPNFAVLAA
MMAR_0434|M.marinum_M YALVAALTTVPLVRWTAHWPRRHALMVSLVCLTISQLISALAPNFAVLAA
MUL_1084|M.ulcerans_Agy99 YALVAALTTVPLVRWTAHWPRRHALMVSLVCLTISQLISALAPNFAVLAA
MAV_4987|M.avium_104 YALVAALTTIPLVRWTAHLPRRRVLVASLTCLTASQLISALAPNFAVLAA
MSMEG_0232|M.smegmatis_MC2_155 YALIAALTTVALVRLTARWPRRRALLVTMVCLTVSQVISAMAPNFAVLAG
TH_3988|M.thermoresistible__bu PGVLAGAVAVLTPTLIGRLDRRLALVSALLLVFVSAATSAIAPNLPVLLA
MLBr_01562|M.leprae_Br4923 YVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCAVAWDEVTLVI
: *: :: : . . .* .*
Mflv_1840|M.gilvum_PYR-GCK ARFLAGLPHGAYFGVAALVAAHLMGPQHRAKAVAHVLTGLTVATVLGVPV
Mvan_4890|M.vanbaalenii_PYR-1 ARFLAGLPHGAYFGVAALVAAHLMGPQNRAKAVAHVLTGLTVATVLGVPI
MAB_1429|M.abscessus_ATCC_1997 SRFVAGLPHGAYFGVAALVAAHLAGPGERAKAVGQTMLGLSVANVVGVPV
Mb0197|M.bovis_AF2122/97 GRVLCAVTHGLLWAVIAPIATRLVPPSHAGRATTSIYIGTSLALVVGSPL
Rv0191|M.tuberculosis_H37Rv GRVLCAVTHGLLWAVIAPIATRLVPPSHAGRATTSIYIGTSLALVVGSPL
MMAR_0434|M.marinum_M GRALCAITHGLLWSVIAPIATRLVPPSHAGRATTSIYIGTSLALVIGSPL
MUL_1084|M.ulcerans_Agy99 GRALCAITHGLLWSVIAPIATRLVPPSHAGRATTSIYIGTSLALVIGSPL
MAV_4987|M.avium_104 GRVLCAITHGLLWSVIAPIATRLVPPSHAGRATMSIYVGTSLALVVGSPL
MSMEG_0232|M.smegmatis_MC2_155 GRVLCALAHGLMWSVIAPIGVRLVPPSHAGRATAAVYVGTGLALVVGNPL
TH_3988|M.thermoresistible__bu SRLFAGMAIGVFWALLAVVVARVAHPDDLPKALTLAFGGVSVAVVLGVPF
MLBr_01562|M.leprae_Br4923 ARLLQGIGSAIASPTGLALVATTFSKGSARNTATAVFAAMTAIGSVMGLV
.* . .: . : . .: . : .
Mflv_1840|M.gilvum_PYR-GCK ASWLGQSLGWR-AAFGLVVAVGVLTLTALALWLPVQLRLMRSTSPLTELG
Mvan_4890|M.vanbaalenii_PYR-1 ASWLGQSLGWR-AAFGLVVGVGLVTLTALWCWLPFQLKFMRATSPLTELG
MAB_1429|M.abscessus_ATCC_1997 ATWLGNTVGWR-SAFAVVAVIGAITIAALLRYLP-GLTDMRITNPLTELG
Mb0197|M.bovis_AF2122/97 TAAMSLMWGWRLAAVCVTGAAAAVALAARLALPEMVLRADQLEHVGRRAR
Rv0191|M.tuberculosis_H37Rv TAAMSLMWGWRLAAVCVTGAAAAVALAARLALPEMVLRADQLEHVGRRAR
MMAR_0434|M.marinum_M TAALSLMWGWRLAAVCVTVAAAVVTVAARLLLPEMVLSADQLQYVGPRSR
MUL_1084|M.ulcerans_Agy99 TAALSLMWGWRLAAVCVTVAAAVVTVAARLLLPEMVLSADQLQHVGPRSR
MAV_4987|M.avium_104 TAALSLMWGWRLAVVCVTVAAAVVTVAARLMLPEMVLTEHQLAHVGPRSR
MSMEG_0232|M.smegmatis_MC2_155 TAALSELWGWRQAVVAVAIAAAAVTLAARVLLPLMPAERARAGSAG-RPV
TH_3988|M.thermoresistible__bu AAWIGAMVGWRWAFVLVGALGLMVAVALTVLLPPVPVAER--TTLGQLGA
MLBr_01562|M.leprae_Br4923 VGGALTEVSWRLAFLVNVPIGLVMIYLARTALHETHKERMKLDAAGAILA
. .** : . :
Mflv_1840|M.gilvum_PYR-GCK ALRRPQVWLALAVG------------------------------------
Mvan_4890|M.vanbaalenii_PYR-1 ALRRPQVWLALLVG------------------------------------
MAB_1429|M.abscessus_ATCC_1997 ALRRVQVWFTLLIG------------------------------------
Mb0197|M.bovis_AF2122/97 HHRNPRLVKVSVLT------------------------------------
Rv0191|M.tuberculosis_H37Rv HHRNPRLVKVSVLT------------------------------------
MMAR_0434|M.marinum_M HHRNRALIIVSLIT------------------------------------
MUL_1084|M.ulcerans_Agy99 HHRNRALIIVSLIT------------------------------------
MAV_4987|M.avium_104 HHRNGRLITVSLLA------------------------------------
MSMEG_0232|M.smegmatis_MC2_155 HGRG--LLTLSLLT------------------------------------
TH_3988|M.thermoresistible__bu AWSIPAARTGVLVT------------------------------------
MLBr_01562|M.leprae_Br4923 TLACTAAVFAFSMGPEKGWISLTTIGSGLVALVAAIAFAVVERTAENPVV
:
Mflv_1840|M.gilvum_PYR-GCK -----------------MIGFGGMFAVYTYIATTMTDVAGMPRALVPVAL
Mvan_4890|M.vanbaalenii_PYR-1 -----------------MIGFGGMFAVYTYITTTMTDVAGMPRGLVPLAL
MAB_1429|M.abscessus_ATCC_1997 -----------------IVGFGGMFAFYTYISTTLTSVSGLPRALVPLAL
Mb0197|M.bovis_AF2122/97 -----------------MIAVTGHFVSYTYIVVIIRDVVGVRGPNLAWLL
Rv0191|M.tuberculosis_H37Rv -----------------MIAVTGHFVSYTYIVVIIRDVVGVRGPNLAWLL
MMAR_0434|M.marinum_M -----------------MVGVTGHFVSYTYIVVIIRQVVGVRGPSLAWLL
MUL_1084|M.ulcerans_Agy99 -----------------MVGVTGHFVSYTYIVVIIRQVVGVRGPSLAWLL
MAV_4987|M.avium_104 -----------------MVAVTGHFVSYTFIVEIIRNVVGVRGPTLAWVL
MSMEG_0232|M.smegmatis_MC2_155 -----------------LIGVTGHFVAYTFIVAIIRDVVGIDGPHLAWLL
TH_3988|M.thermoresistible__bu -----------------LVMVTAHFTAYTYASPVLQDRIHITESGVGPML
MLBr_01562|M.leprae_Br4923 PFDLFRDRNRLVTFIAIFLAGGVIFSLTVSIGLYIQDILGYSALRAGIGF
:: * . : . :
Mflv_1840|M.gilvum_PYR-GCK MVFGLGMFFGNLVGGRLADRSVVRALYLSMGALCAMLTLFVAASHNPWTA
Mvan_4890|M.vanbaalenii_PYR-1 MMFGLGMVLGNLVGGRLADGSVVRALYLSLGALCGALALFVVASHNPWTA
MAB_1429|M.abscessus_ATCC_1997 SLYGIGMVTGNVLGGWMADRSVVRAIAVGLVLVAAMLLIFAAVSRNPWAA
Mb0197|M.bovis_AF2122/97 AAYGVAGLVSVPLVARPLDRWPKGAVIVGMTGLTAAFTLLTALAFGERHT
Rv0191|M.tuberculosis_H37Rv AAYGVAGLVSVPLVARPLDRWPKGAVIVGMTGLTAAFTLLTALAFGERHT
MMAR_0434|M.marinum_M AAYGVAGVVAVALVARPLDRRPKGTIIFCVAGLTFAFVLLTALAFGGHLA
MUL_1084|M.ulcerans_Agy99 AAYGVAGVVAVALVARPLDRRPKGTIIFCVAGLTFAFVLLTALAFGGHLA
MAV_4987|M.avium_104 AAYGLAGLLSVPLVARPLDHRPKSAVILCMTGLTAAFAVLTALAFGGPTG
MSMEG_0232|M.smegmatis_MC2_155 AGYGVAGLAAMAAMARPLDLWPKASVTAALVVLAASLVTLAVLSAQPRAG
TH_3988|M.thermoresistible__bu FVFGIAGFLGNFAAGPLLRRSMRATYVVLPIGVMVGVXXXGPNSR-----
MLBr_01562|M.leprae_Br4923 IPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLLMVAMLYGWAFMNRGVP
: :. . . :
Mflv_1840|M.gilvum_PYR-GCK LLA----------LFGIGLTGSAVGPALQTRLMDVAHDAQTLAAALNHSA
Mvan_4890|M.vanbaalenii_PYR-1 LLV----------LFLIGLTGSAVGPALQTRLMDVAHDAQTLAAALNHSA
MAB_1429|M.abscessus_ATCC_1997 LAL----------VFAVGVSGTSLVPALQTRLMDVADDAQTLAAALNHSA
Mb0197|M.bovis_AF2122/97 AAT--ALLG-TGAIVLWGALATAVSPMLQSAAMRSGGDDPDGASGLYVTA
Rv0191|M.tuberculosis_H37Rv AAT--ALLG-TGAIVLWGALATAVSPMLQSAAMRSGGDDPDGASGLYVTA
MMAR_0434|M.marinum_M PMT--ALVVGTGAIVLWGAAATAVSPMLQSAAMRSGADDPDGASGLYVTA
MUL_1084|M.ulcerans_Agy99 PMT--ALVVGTGAIVLWGAAVTAVSPMLQSAAMRSGADDPDGASGLYVTA
MAV_4987|M.avium_104 ATT--ALIG-TAAIVLWGAMATAVSPMMQSAAMRNGADDPDGASGLYVTA
MSMEG_0232|M.smegmatis_MC2_155 VTTGAVMVIGTVAIVLWGASSTALPPMLQTSAMRTSPEDPDGASGRYVAA
TH_3988|M.thermoresistible__bu ----------------WRPDGTRM--------------------------
MLBr_01562|M.leprae_Br4923 YFPN----LLAPIVVGGIGIGMAVVPLTLSAIAGVGFDRIGPVSAIALML
:
Mflv_1840|M.gilvum_PYR-GCK LNIG---------------------------NATGAWVGGLVIAAGYGYT
Mvan_4890|M.vanbaalenii_PYR-1 LNIG---------------------------NATGAWVGGLVIAAGLGYT
MAB_1429|M.abscessus_ATCC_1997 LNIA---------------------------NAGGAWLGGLVITAGYGYR
Mb0197|M.bovis_AF2122/97 FQIG---------------------------IMAGALLGGLLYERS-LAM
Rv0191|M.tuberculosis_H37Rv FQIG---------------------------IMAGALLGGLLYERS-LAM
MMAR_0434|M.marinum_M FQVG---------------------------IMAGSLIGGLLYERS-VAM
MUL_1084|M.ulcerans_Agy99 FQVG---------------------------IMAGSLIGGLLYERS-VAM
MAV_4987|M.avium_104 FQVG---------------------------IMAGSLLGGLLYERS-VIL
MSMEG_0232|M.smegmatis_MC2_155 FQVG---------------------------IMTGALVGAGVYEAVGLTA
TH_3988|M.thermoresistible__bu --------------------------------------------------
MLBr_01562|M.leprae_Br4923 QSLGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDAQLRALDHGYTYGLL
Mflv_1840|M.gilvum_PYR-GCK APAGAGAVLALAGLVVLTVSVVLQKR------------
Mvan_4890|M.vanbaalenii_PYR-1 APAAAGAVLALAGLAVLTVSVLLQKRG-----------
MAB_1429|M.abscessus_ATCC_1997 APSLVGAALAIAGLVVLGLSVLYARSRAVALPSRAG--
Mb0197|M.bovis_AF2122/97 MLTASAGLMGVALFGMTVSQHLFENPTLSPGDG-----
Rv0191|M.tuberculosis_H37Rv MLTASAGLMGVALFGMTVSQHLFENPTLSPGDG-----
MMAR_0434|M.marinum_M MLTASAGLMGVALVGMTSARRVFEAPVAVAGATSHNQ-
MUL_1084|M.ulcerans_Agy99 MLTASAGLMGVALVGMTSARRVFEAPVAVAGATSHNQ-
MAV_4987|M.avium_104 MLSASGVLMAVALVGIAANRRMLDVAPTSSRDS-----
MSMEG_0232|M.smegmatis_MC2_155 MIGTAAVLITAAMCGALAVPDVFNRSRDLSTR------
TH_3988|M.thermoresistible__bu --------------------------------------
MLBr_01562|M.leprae_Br4923 WVAGAVVIVGVAALFIGYTPEQVAYAQEVKEAVDAGEL