For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LDRPNPGALHGSLLTALGTGIVSGQYPPGHVLTLEAVSTEHGVSRSVAREAVRVLESMGMLESRRRVGIT VQPAESWNVFDPVVIRWRLEVGDRTAQLVSLSELRLGFEPAAAALAARRASPHQCRIMATAVSDMVVHGR TGDLDAYLLADKLFHQTLLEASGNEMFRALNGVVAEVLTGRTHHGLMPEKPNIAAIALHDEVARAIRMGE EQQAERAMRAIIDESASAIVEDSPPSD*
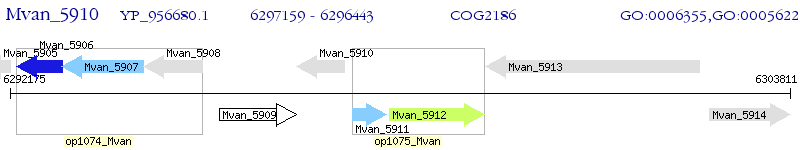
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5910 | - | - | 100% (238) | GntR domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_4590 | - | 7e-10 | 27.52% (218) | GntR domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_0952 | - | 1e-09 | 26.58% (222) | GntR domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0505 | - | 9e-09 | 26.34% (186) | GntR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0501 | - | 1e-114 | 88.41% (233) | GntR domain-containing protein |
| M. tuberculosis H37Rv | Rv0494 | - | 9e-09 | 26.34% (186) | GntR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1705c | - | 6e-10 | 29.41% (221) | GntR family transcriptional regulator |
| M. marinum M | MMAR_0820 | - | 4e-10 | 23.61% (233) | GntR family transcriptional regulator |
| M. avium 104 | MAV_2542 | - | 3e-10 | 34.38% (160) | regulatory protein GntR, HTH |
| M. smegmatis MC2 155 | MSMEG_0454 | - | 1e-112 | 85.59% (236) | GntR-family protein transcriptional regulator |
| M. thermoresistible (build 8) | TH_4024 | - | 2e-08 | 28.37% (215) | PUTATIVE transcriptional regulator, GntR family |
| M. ulcerans Agy99 | MUL_4564 | - | 4e-10 | 23.61% (233) | GntR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0505|M.bovis_AF2122/97 -----MVEPMNQSSVFQPPDRQRVDERIATTIADAILDGVFPPGSTLPPE
Rv0494|M.tuberculosis_H37Rv -----MVEPMNQSSVFQPPDRQRVDERIATTIADAILDGVFPPGSTLPPE
MMAR_0820|M.marinum_M ---------MNQSSILQPPDRQRVDEQIATSIADAILDGVFPPGSALPPE
MUL_4564|M.ulcerans_Agy99 ---------MNQSSILQPPDRQRVDEQIATSIADAILDGVFPPGSALPPE
Mvan_5910|M.vanbaalenii_PYR-1 --------------MLDRPNPGALHGSLLTALGTGIVSGQYPPGHVLTLE
Mflv_0501|M.gilvum_PYR-GCK --------------MLERPVVGALHSSLVTALGTGIVSGKYPAGCVLTLE
MSMEG_0454|M.smegmatis_MC2_155 --------------MLDRPIAGELHGSLVAALGTGIVSGRYQPGHVLNLE
MAB_1705c|M.abscessus_ATCC_199 ---------MPVPPSEGIAARSLLRDNAYKALRTAIVDGTLAPGERLRDD
MAV_2542|M.avium_104 MPKREPLAPMATSQNNGAVRSPKTAEIVADTLRRMIVEGQLKDGDFLPYE
TH_4024|M.thermoresistible__bu --------------MVRRVRAPRVAEVVASAIRDDILSGRLTEGDVLPTQ
:: *:.* * * :
Mb0505|M.bovis_AF2122/97 RDLAERLGVNRTSLRQGLARLQQMGLIEVRHGSGSVVRDPE--GLTHPAV
Rv0494|M.tuberculosis_H37Rv RDLAERLGVNRTSLRQGLARLQQMGLIEVRHGSGSVVRDPE--GLTHPAV
MMAR_0820|M.marinum_M RELAEQLGVNRTSLRQGLARLQQMGLIETRQGSGNVVRDPQ--SLTHPAV
MUL_4564|M.ulcerans_Agy99 RELAEQLGVNRTSLRQGLARLQQMGLIETRQGSGNVVRDPQ--SLTHPAV
Mvan_5910|M.vanbaalenii_PYR-1 A-VSTEHGVSRSVAREAVRVLESMGMLESRRRVGITVQPAESWNVFDPVV
Mflv_0501|M.gilvum_PYR-GCK A-VSAEHGVSRSVAREAVRVLESMGMVESRRRVGITVLPADRWNLFDPVV
MSMEG_0454|M.smegmatis_MC2_155 G-ISAEHGVSRSVAREAVRVLESMGMVESRRRVGITVQDPEKWNVFDPVL
MAB_1705c|M.abscessus_ATCC_199 E-LSRWLGVSRTPIREALARLEQAGLVQTQPGRHTIVSPID---------
MAV_2542|M.avium_104 AELMDHFQVSRPSLREAVRVLESDRLVEVRRGSRTGARVRVPGPEIVARP
TH_4024|M.thermoresistible__bu EHLFTEFGVSPPALREAIRILETEGLVSVRRGNVGGAVVHLPSARRTAQM
: *. . *:.: *: ::. : .
Mb0505|M.bovis_AF2122/97 VEALVRKLG-PDFLVELLEIRAALGPLIGRLAAARSTPEDAEALCAALEV
Rv0494|M.tuberculosis_H37Rv VEALVRKLG-PDFLVELLEIRAALGPLIGRLAAARSTPEDAEALCAALEV
MMAR_0820|M.marinum_M VEALVRKLG-PDFLVEVLEIRAALGPLIGRLAAERNSSEDAEALRAALAE
MUL_4564|M.ulcerans_Agy99 VEALVRKLG-PDFLVEVLEIRAALGPLIGRLAAERNSSEDAEALRAALAE
Mvan_5910|M.vanbaalenii_PYR-1 IRWRLEVGDRTAQLVSLSELRLGFEPAAAALAARRASPHQCRIMATAVSD
Mflv_0501|M.gilvum_PYR-GCK IRWRLEVGDRTAQLVSLSELRMGFEPAAAALAARRASPHQCRVMATAVSD
MSMEG_0454|M.smegmatis_MC2_155 IRWRLQAGDRTAQLVSLSELRLGFEPAAAALAARRASPHQCRVLATAVSD
MAB_1705c|M.abscessus_ATCC_199 ----------VREARAAQSVAAAMHELAVREAITNISPGDLAAMRHANTR
MAV_2542|M.avium_104 AGFLLEMAG--TTLADVMTARMGIEPYAARLLAEDGTAAAHRELRELIEE
TH_4024|M.thermoresistible__bu IAMVLQSRS--ATPADVSEALLHLEPICAGMCAARPDRMSEVVPYLQDEI
:
Mb0505|M.bovis_AF2122/97 VQQADTAA---ARQAADLAYFRVLIHSTRNRALGLLYRWVEHAFGGREHA
Rv0494|M.tuberculosis_H37Rv VQQADTAA---ARQAADLAYFRVLIHSTRNRALGLLYRWVEHAFGGREHA
MMAR_0820|M.marinum_M VADADTAP---ARQAADLAYFRVLIHGTRNRALGLLYRWVEQAFGGREHE
MUL_4564|M.ulcerans_Agy99 VADADTAP---ARQAADLAYFRVLIHGTRNRALGLLYRWVEQAFGGREHD
Mvan_5910|M.vanbaalenii_PYR-1 MVVHGRTGDLDAYLLADKLFHQTLLEASGNEMFRALNGVVAEVLTGRTHH
Mflv_0501|M.gilvum_PYR-GCK MVMHGRTGDLDAYLLADKLFHQTMLEASGNEMFRALNGVVAEVLAGRTHH
MSMEG_0454|M.smegmatis_MC2_155 MVMHGRNGDLAAYLAADKLFHQTMLEASGNEMFRALNGAVAEVLAGRTHH
MAB_1705c|M.abscessus_ATCC_199 FAAALDAHDPDAALAADDEFHAVAVEASANPYIAAVLEQVTPVLR-RLER
MAV_2542|M.avium_104 IPAAWETG---KLGAASTALHRRLVELSGNATLTVIVGMLHEIFERHMTA
TH_4024|M.thermoresistible__bu DLQSTSFADTARFVSNARRFHEMLVARCGNEPMILLIGALELIWSAHEAG
. : * : : : :
Mb0505|M.bovis_AF2122/97 LTGAYD---------DADPVLTDLRAINGAVLAGDPAAAAATVEAYLNAS
Rv0494|M.tuberculosis_H37Rv LTGAYD---------DADPVLTDLRAINGAVLAGDPAAAAATVEAYLNAS
MMAR_0820|M.marinum_M LTGAYE---------DADPVIADLTAINDAVLAGDPETAAAAVEGYLNAS
MUL_4564|M.ulcerans_Agy99 LTGAYE---------DADPVIADLTAINDAVLAGDPETAAAAVEGYLNAS
Mvan_5910|M.vanbaalenii_PYR-1 GLMPEK---------PNIAAIALHDEVARAIRMGEEQQAERAMRAIIDES
Mflv_0501|M.gilvum_PYR-GCK GMMPEK---------PNMAAIALHDEVARAIRMGEQEQAQRAMWAIIDES
MSMEG_0454|M.smegmatis_MC2_155 GLMPEK---------PNIDAIALHDEVARAIRMGEADQAERAMRAIIDES
MAB_1705c|M.abscessus_ATCC_199 IRFASR---------DGRHSVALHEQIIARCEAGDSEGAGRAARDNWHTL
MAV_2542|M.avium_104 AFLSVQN---VVPKAQYNKLMRSYTRLADLVAARDGAEAEAHWRRHMENA
TH_4024|M.thermoresistible__bu VWSDDSPGGGRPEAATMRAALRDHRRILDAVIDGDSARAVRLAQKHLAAA
: : : *
Mb0505|M.bovis_AF2122/97 ALRMVKSYRDRA-----------
Rv0494|M.tuberculosis_H37Rv ALRMVKSYRDRA-----------
MMAR_0820|M.marinum_M ALRMIMCYRAAYAGEDPER----
MUL_4564|M.ulcerans_Agy99 ALRMIMCYRAAYAGEDPER----
Mvan_5910|M.vanbaalenii_PYR-1 ASAIVEDSPPSD-----------
Mflv_0501|M.gilvum_PYR-GCK AMAIVEDGGAK------------
MSMEG_0454|M.smegmatis_MC2_155 VSAIVEVGPPGAGTAAQA-----
MAB_1705c|M.abscessus_ATCC_199 IAAPEGAAHEERGAG--------
MAV_2542|M.avium_104 SAELLKGHEKTKVRDIMH-----
TH_4024|M.thermoresistible__bu RRHTLACGADTTIEAKLVSRPDH