For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTRNTPLAPMIGPDAVASRAAVRSPKTAELVAGTLRRMVVEGQLKDGDFLPNEAELMAHFGVSRPTLREA VRVLESERLVEVRRGSRTGARVRVPGPEIVARPAGLLLELSGATIADLLTAKSGIEPMAARLLAESGSPE AFDELERMLEELAADSHKSSRLAEATGLFHLRVVQLSGNATLSIVAGMLHEITVRHTAFVFNERRPVSKA DYDTLLRSYRKLMQFLRAGDAVGAEAHWRKHLDTTRELLLTGLESVPVRDVMG
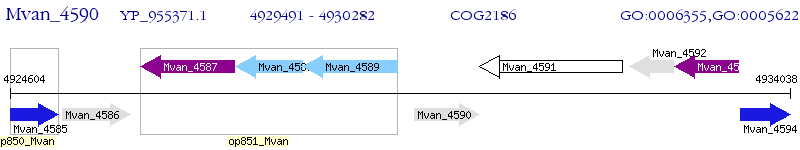
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4590 | - | - | 100% (263) | GntR domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_1986 | - | 1e-27 | 32.03% (231) | regulatory protein GntR, HTH |
| M. vanbaalenii PYR-1 | Mvan_1846 | - | 2e-17 | 31.64% (177) | regulatory protein GntR, HTH |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3086c | - | 1e-14 | 27.56% (225) | GntR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_2114 | - | 1e-132 | 91.63% (263) | GntR domain-containing protein |
| M. tuberculosis H37Rv | Rv3060c | - | 1e-14 | 27.56% (225) | GntR family transcriptional regulator |
| M. leprae Br4923 | MLBr_01188 | - | 1e-09 | 36.05% (86) | hypothetical protein MLBr_01188 |
| M. abscessus ATCC 19977 | MAB_3018 | - | 2e-11 | 31.14% (167) | GntR family transcriptional regulator |
| M. marinum M | MMAR_3975 | - | 2e-29 | 34.52% (252) | FadR family transcriptional regulator |
| M. avium 104 | MAV_2542 | - | 4e-81 | 60.08% (263) | regulatory protein GntR, HTH |
| M. smegmatis MC2 155 | MSMEG_5201 | - | 1e-114 | 80.77% (260) | regulatory protein GntR, HTH |
| M. thermoresistible (build 8) | TH_0492 | - | 3e-60 | 68.29% (164) | regulatory protein GntR, HTH |
| M. ulcerans Agy99 | MUL_3832 | - | 1e-29 | 34.80% (250) | FadR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3086c|M.bovis_AF2122/97 --------------MSTEPDAVWTDKRASKIARRIEADIVRRGWPIGASL
Rv3060c|M.tuberculosis_H37Rv --------------MSTEPDAVWTDKRASKIARRIEADIVRRGWPIGASL
Mvan_4590|M.vanbaalenii_PYR-1 MTRNTPLAPMIGPDAVASRAAVRSPKTAELVAGTLRRMVVEGQLKDGDFL
Mflv_2114|M.gilvum_PYR-GCK MARSTPLAPMIGPDAVASRTAVRSPKTAELVAGTLRRMVVEGQLKDGDFL
MSMEG_5201|M.smegmatis_MC2_155 MAGSTPLAPMIGPDAITPRAAVRSPKTAELVAGTLRRMVVEGQLKDGDFL
TH_0492|M.thermoresistible__bu --------------------------------------------------
MAV_2542|M.avium_104 MPKREPLAPMATS---QNNGAVRSPKTAEIVADTLRRMIVEGQLKDGDFL
MMAR_3975|M.marinum_M --------MTALGIGPDPRRRLSAPRIAEIVADELRRQIIDGELADGDLL
MUL_3832|M.ulcerans_Agy99 --------MTALGIGPDPRRRLSAPRIAEIVADELRRQIIDGELADGDLL
MLBr_01188|M.leprae_Br4923 -------------------------------------------------M
MAB_3018|M.abscessus_ATCC_1997 ----------------MALQPIARQSIPDEIFSQLAAQVLTGDRIPGDTL
Mb3086c|M.bovis_AF2122/97 GSESALQQRFCVSRSVLREAVRLVEHHQVARMRRGPNGGLFICEPNAGPA
Rv3060c|M.tuberculosis_H37Rv GSESALQQRFCVSRSVLREAVRLVEHHQVARMRRGPNGGLFICEPNAGPA
Mvan_4590|M.vanbaalenii_PYR-1 PNEAELMAHFGVSRPTLREAVRVLESERLVEVRRGSRTGARVRVPGPEIV
Mflv_2114|M.gilvum_PYR-GCK PNEAELMAHFGVSRPTLREAVRVLESERLVEVRRGSRTGARVRVPGPEIV
MSMEG_5201|M.smegmatis_MC2_155 PNEAELMSHFGVSRPTLREAVRVLESERLVEVRRGSRTGARVRVPGPEIV
TH_0492|M.thermoresistible__bu -------------------------------------------------V
MAV_2542|M.avium_104 PYEAELMDHFQVSRPSLREAVRVLESDRLVEVRRGSRTGARVRVPGPEIV
MMAR_3975|M.marinum_M PRQEVLVEQFNVSLVSLREALRILETEGLVSVRRGNRGGAVVHAPAKTSA
MUL_3832|M.ulcerans_Agy99 PRQEVLVEQFNVSLVSLREALRILETEGLVSVRRGNRGGAVVHAPAKASA
MLBr_01188|M.leprae_Br4923 PRQEALVEPLNVSLLSFREALQIMDTERLISLRRGNRGSVVVHTPTRTSA
MAB_3018|M.abscessus_ATCC_1997 PSERALAEALGVSRAAVREALGRLERSGLIQIRQGGSTVIRDYRSDAGFD
Mb3086c|M.bovis_AF2122/97 TRAVVIYLE--YLGTTIGDLLGARLVLEPLAASLAAEHIDEPG-----IE
Rv3060c|M.tuberculosis_H37Rv TRAVVIYLE--YLGTTIGDLLGARLVLEPLAASLAAEHIDEPG-----IE
Mvan_4590|M.vanbaalenii_PYR-1 ARPAGLLLE--LSGATIADLLTAKSGIEPMAARLLAE--SGSPEAFDELE
Mflv_2114|M.gilvum_PYR-GCK ARPAGLLLE--LSGATIADLLTAKSAIEPMAARLLAE--SGSAAAFDELE
MSMEG_5201|M.smegmatis_MC2_155 ARPVGLLLE--LSGADIADLLVARSAIEPMAARLLAE--NGTEEQFAELD
TH_0492|M.thermoresistible__bu ARPAALLLE--LSGATVADVMVARSGTEPMAARLLAE--SGNTEAFDELE
MAV_2542|M.avium_104 ARPAGFLLE--MAGTTLADVMTARMGIEPYAARLLAE--DGTAAAHRELR
MMAR_3975|M.marinum_M AYMLGLLLQ--SESVAVSDLGAALQELEPACAALAAQRPDRADTLVPELK
MUL_3832|M.ulcerans_Agy99 AYMLGLLLQ--SESVAVSDLGAALQELEPACAALAAQRPDRADTLVPELK
MLBr_01188|M.leprae_Br4923 AYMLGLLLQ--SKSIALADLGAALQELEPACAALAAQ-------------
MAB_3018|M.abscessus_ATCC_1997 VLPLLLAYGGDVDRRTLASVIEARAIIGPQVARLAAQRAHQTPGVADRLR
. : :..: * * * * *:
Mb3086c|M.bovis_AF2122/97 RLRAVLRAEERWRPGLPPPPEQFYRVLAEQSKNPVLQLFIDILMRLTKRY
Rv3060c|M.tuberculosis_H37Rv RLRAVLRAEERWRPGLPPPPEQFYRVLAEQSKNPVLQLFIDILMRLTKRY
Mvan_4590|M.vanbaalenii_PYR-1 RMLEELAADSHKSSRLAEATGLFHLRVVQLSGNATLSIVAGMLHEITVRH
Mflv_2114|M.gilvum_PYR-GCK QLLEELVADSHQSPRLAEATGAFHLRVVQLSGNATLSIVAGMLHEITVRH
MSMEG_5201|M.smegmatis_MC2_155 RMLEEHIPSDWQSDRLAETTGDFHRRVVELSGNATLGIIAGMLHEITVRH
TH_0492|M.thermoresistible__bu KMLTEHIPAGYESGRLAETTGDFHLRVVQLSGNTTLSIIAGMLHEITVRH
MAV_2542|M.avium_104 ELIEE-IPAAWETGKLGAASTALHRRLVELSGNATLTVIVGMLHEIFERH
MMAR_3975|M.marinum_M RVNEAMSEHLDDGAMFTEIGRQFHDLIVRGCGNHTIIAVVGSLETLWTSH
MUL_3832|M.ulcerans_Agy99 RVNEAMSEHLDDGAMFTEIGRQFHDLIVRGCGNHTIIAVVGSLETLWTSH
MLBr_01188|M.leprae_Br4923 --------------------------------------------------
MAB_3018|M.abscessus_ATCC_1997 AMTSELEAEDDPLQRMWQALG-FWELVIDTADSIAFRLVFNTMRDSYVRA
Mb3086c|M.bovis_AF2122/97 VQKSGTQSAG----EAVEAAGQVHNEHSDIVAAVTAGDSAWAKTLSERHV
Rv3060c|M.tuberculosis_H37Rv VQKSGTQSAG----EAVEAAGQVHNEHSDIVAAVTAGDSAWAKTLSERHV
Mvan_4590|M.vanbaalenii_PYR-1 TAFVFNERR----PVSKADYDTLLRSYRKLMQFLRAGDAVGAEAHWRKHL
Mflv_2114|M.gilvum_PYR-GCK TAFVFNERR----PVSKADYETLLRSYRRLLQFLRSRDADGAEAHWRRHL
MSMEG_5201|M.smegmatis_MC2_155 HQFLFREHR----PVSKSDYDKLMRSYRKLMQIMRSGDGDAAEAHWRTHL
TH_0492|M.thermoresistible__bu TEVALRGER----SLSKTDYDKLIRSYRRLLKLVRAGDGAAAEAHWRTHL
MAV_2542|M.avium_104 MTAAFLSVQN---VVPKAQYNKLMRSYTRLADLVAARDGAEAEAHWRRHM
MMAR_3975|M.marinum_M EQQWADESAARGTYPSLTKRRAVLNTHIKLTETIAEGDVDRARRIAGRHL
MUL_3832|M.ulcerans_Agy99 EQQWADESAARGTYPSLAKRRAVLNAHIKLTETIAEGDVDRARRIAGRHL
MLBr_01188|M.leprae_Br4923 --------------------------------------------------
MAB_3018|M.abscessus_ATCC_1997 LDVLVN---------VMAAEVGDIGHYRALADAIALADPDAAQDAAAGML
Mb3086c|M.bovis_AF2122/97 EAVAGWLQQHQRGNDAAVRNGGRAREPRRAQQLILGAPRGKLAEVLAATI
Rv3060c|M.tuberculosis_H37Rv EAVAGWLQQHQRGNDAAVRNGGRAREPRRAQQLILGAPRGKLAEVLAATI
Mvan_4590|M.vanbaalenii_PYR-1 DTTRELLLTGLESVPVRDVMG-----------------------------
Mflv_2114|M.gilvum_PYR-GCK DKTRELLLAGLESVPVRDVMG-----------------------------
MSMEG_5201|M.smegmatis_MC2_155 DTARALMLQGLESVKVRDVMG-----------------------------
TH_0492|M.thermoresistible__bu DTARQLMLEGLETIRVRDIMG-----------------------------
MAV_2542|M.avium_104 ENASAELLKGHEKTKVRDIMH-----------------------------
MMAR_3975|M.marinum_M ADTQTYVLSGRAEQRIYALSPQALSRPRDMRRP-----------------
MUL_3832|M.ulcerans_Agy99 ADTQTYVLSGRAEQRIYALSPQALSRPRDMRRP-----------------
MLBr_01188|M.leprae_Br4923 --------------------------------------------------
MAB_3018|M.abscessus_ATCC_1997 ALGTKAFDKLLREMEKER--------------------------------
Mb3086c|M.bovis_AF2122/97 GDDIAASGWQVGSVFGTETALLERYQVSRAVLREAVRLLEYHAIAHMRRG
Rv3060c|M.tuberculosis_H37Rv GDDIAASGWQVGSVFGTETALLERYQVSRAVLREAVRLLEYHAIAHMRRG
Mvan_4590|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2114|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5201|M.smegmatis_MC2_155 --------------------------------------------------
TH_0492|M.thermoresistible__bu --------------------------------------------------
MAV_2542|M.avium_104 --------------------------------------------------
MMAR_3975|M.marinum_M --------------------------------------------------
MUL_3832|M.ulcerans_Agy99 --------------------------------------------------
MLBr_01188|M.leprae_Br4923 --------------------------------------------------
MAB_3018|M.abscessus_ATCC_1997 --------------------------------------------------
Mb3086c|M.bovis_AF2122/97 PGGGLVVTTPQPQASIDTIALYLQYRKPSREDLRCVRDAIEIDNVAKVVK
Rv3060c|M.tuberculosis_H37Rv PGGGLVVTTPQPQASIDTIALYLQYRKPSREDLRCVRDAIEIDNVAKVVK
Mvan_4590|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2114|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5201|M.smegmatis_MC2_155 --------------------------------------------------
TH_0492|M.thermoresistible__bu --------------------------------------------------
MAV_2542|M.avium_104 --------------------------------------------------
MMAR_3975|M.marinum_M --------------------------------------------------
MUL_3832|M.ulcerans_Agy99 --------------------------------------------------
MLBr_01188|M.leprae_Br4923 --------------------------------------------------
MAB_3018|M.abscessus_ATCC_1997 --------------------------------------------------
Mb3086c|M.bovis_AF2122/97 RRSEPEVASFLDTLGRPRLDNPTDDVRAAAVEEFRFHVGLARAAGNTMLD
Rv3060c|M.tuberculosis_H37Rv RRSEPEVASFLDTLGRPRLDNPTDDVRAAAVEEFRFHVGLARAAGNTMLD
Mvan_4590|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2114|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5201|M.smegmatis_MC2_155 --------------------------------------------------
TH_0492|M.thermoresistible__bu --------------------------------------------------
MAV_2542|M.avium_104 --------------------------------------------------
MMAR_3975|M.marinum_M --------------------------------------------------
MUL_3832|M.ulcerans_Agy99 --------------------------------------------------
MLBr_01188|M.leprae_Br4923 --------------------------------------------------
MAB_3018|M.abscessus_ATCC_1997 --------------------------------------------------
Mb3086c|M.bovis_AF2122/97 LFLLILVELFRRHLSSTEQALPTWSDVVAVGHAHVRILEAIGSGDDSLAR
Rv3060c|M.tuberculosis_H37Rv LFLLILVELFRRHLSSTEQALPTWSDVVAVGHAHVRILEAIGSGDDSLAR
Mvan_4590|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2114|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5201|M.smegmatis_MC2_155 --------------------------------------------------
TH_0492|M.thermoresistible__bu --------------------------------------------------
MAV_2542|M.avium_104 --------------------------------------------------
MMAR_3975|M.marinum_M --------------------------------------------------
MUL_3832|M.ulcerans_Agy99 --------------------------------------------------
MLBr_01188|M.leprae_Br4923 --------------------------------------------------
MAB_3018|M.abscessus_ATCC_1997 --------------------------------------------------
Mb3086c|M.bovis_AF2122/97 CRTRRHLDAAASWWL
Rv3060c|M.tuberculosis_H37Rv CRTRRHLDAAASWWL
Mvan_4590|M.vanbaalenii_PYR-1 ---------------
Mflv_2114|M.gilvum_PYR-GCK ---------------
MSMEG_5201|M.smegmatis_MC2_155 ---------------
TH_0492|M.thermoresistible__bu ---------------
MAV_2542|M.avium_104 ---------------
MMAR_3975|M.marinum_M ---------------
MUL_3832|M.ulcerans_Agy99 ---------------
MLBr_01188|M.leprae_Br4923 ---------------
MAB_3018|M.abscessus_ATCC_1997 ---------------