For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGPAITERALCYRCPVPRPRVIDLDHLMDAVEQLAADAGPRAVTIRALSQRTAISNGAIYHAFGSRGGLL GQVWLRAARRFLSEQRSAVARALADGVTPETAVEAVVAAAQCPATFLDGHPATAMYLLALPRNELLGARD VSDELADDLRGLDGELAGLFIELAEHMWRRRDRHAVAAIRACVVDLPTALLLRGQHPPDPAARDRLTAAV RAVLALTPPTSRNTPAH
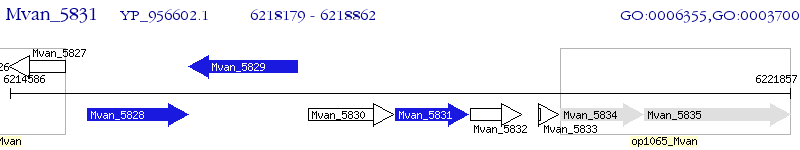
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5831 | - | - | 100% (227) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1548 | - | 1e-14 | 28.65% (178) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_2880 | - | 2e-05 | 32.41% (108) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4881 | - | 3e-15 | 30.54% (203) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_02497 | - | 4e-27 | 44.59% (157) | hypothetical protein MLBr_02497 |
| M. abscessus ATCC 19977 | MAB_2466c | - | 4e-57 | 54.90% (204) | putative transcriptional regulator, TetR |
| M. marinum M | MMAR_1142 | - | 9e-05 | 27.03% (111) | transcriptional regulator |
| M. avium 104 | MAV_1113 | - | 4e-73 | 68.29% (205) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_6556 | - | 3e-36 | 42.36% (203) | putative transcriptional regulator |
| M. thermoresistible (build 8) | TH_0467 | - | 6e-05 | 31.58% (76) | POSSIBLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY |
| M. ulcerans Agy99 | MUL_0905 | - | 6e-05 | 27.03% (111) | transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5831|M.vanbaalenii_PYR-1 MGPAITERALCYRCPVPRPRVIDLDHLMDAVEQLAADAGPRAVTIRALSQ
MAV_1113|M.avium_104 --------------------MLDFDHLMDVAEQLAAESGPPAITIRALSE
MLBr_02497|M.leprae_Br4923 --------------------------------------------------
MAB_2466c|M.abscessus_ATCC_199 ---------------MPRPRIHSVEDLLDATERIAVQDGPAAVTVRAVSQ
MSMEG_6556|M.smegmatis_MC2_155 ---------------MPRPRMHDPQAILDVAESLAVRCGPAAVTIRAVGD
MMAR_1142|M.marinum_M --------------------------MLEAAADLFAERGPAATSIRDIAA
MUL_0905|M.ulcerans_Agy99 ----MTTGSAIRSRKTPTGREEVAAAVLEAAADLFAERGPAATSIRDIAA
Mflv_4881|M.gilvum_PYR-GCK ---------------MPRPARYTVDVLLDAAAELLAAEGPAAVTMSAVAR
TH_0467|M.thermoresistible__bu ----VVSRRDKRVPYAEASRVLLRDSILDGMRELLQTRDWSQITLADVAK
Mvan_5831|M.vanbaalenii_PYR-1 RTAISNGAIYHAFGSRGGLLGQVWLRAARRFLSEQRSAVARALADGVTPE
MAV_1113|M.avium_104 ATAISNGAIYHAFGSRGGLLGHVWLRAGKRYLSFQRAAVAKALSRGVSGE
MLBr_02497|M.leprae_Br4923 -----------MFASRAALLGRVWLRAGYRFLKTQTALVDQALSASVRR-
MAB_2466c|M.abscessus_ATCC_199 ATGISNGAIYHAFGSRGGMVGHAWLRAAQRFLDMQRDAVDGALAESETRT
MSMEG_6556|M.smegmatis_MC2_155 AVGVSNGALYHEFGSRSGLLTRAWLRAVHRLQTVLTELVETAEPG-----
MMAR_1142|M.marinum_M RSKVNHGLVFRHFGAKEQLVGAVLDYLGTHLTELLR--------------
MUL_0905|M.ulcerans_Agy99 RSKVNHGLVFRHFGAKEQLVGAVLDYLGTHLTELLR--------------
Mflv_4881|M.gilvum_PYR-GCK STGAPSGSVYHRFPTRAALCGELWVRTEERFQEELLEALSGDGDP-----
TH_0467|M.thermoresistible__bu AAGISRQTIYNEFGSRQGLAQGYALRLADRLVDEVDEAIANNVGD-----
* :: : :
Mvan_5831|M.vanbaalenii_PYR-1 TAVEAVVAAAQCPATFLDGHPATAMYLLALPRNELLGARDVSDELADDLR
MAV_1113|M.avium_104 AAVDAVVAAAECPATFSHEHPASARFLLRVRRDELLGAADVPAELADELR
MLBr_02497|M.leprae_Br4923 AGVEAVVAAADAPVVFAEQYPDSSRLLLTVRREELLKS-FLPNEIEIELR
MAB_2466c|M.abscessus_ATCC_199 AAVNAVVAAADTPAAFAERFPESSRLVLSVRREDVLGS-GIPEEVARDMA
MSMEG_6556|M.smegmatis_MC2_155 --AAAIAAAAEASIVLAERHPSSAALVLQTRRDELIGA-----DPPAEVK
MMAR_1142|M.marinum_M ----AGTPADDLERALDRQMRVWARTLLDGYPAEQLQT---------RFP
MUL_0905|M.ulcerans_Agy99 ----AGTPADDLERALDRQMRVWARTLLDGYPAEQLQT---------RFP
Mflv_4881|M.gilvum_PYR-GCK --QNRCVAGALRNMQWCRENPVEAHVLLAGP--DQLDMGEWPDTVAGRRK
TH_0467|M.thermoresistible__bu -IHAAFLAGFRKFFAESAADPLVISLLTGSAKPDLLQL------ITTDSA
.. : : :
Mvan_5831|M.vanbaalenii_PYR-1 GLDGELAGLFIELAEHMWRRRDRHAVAAIRACVVDLPTALLLRGQHPPDP
MAV_1113|M.avium_104 RLDDDLAHLFIELAEKVWGRRDQQAVGAIRTCVVELPTALLIDAHALHDA
MLBr_02497|M.leprae_Br4923 ELDALLISLLTRLAVVIWDRKDTGAIDTITTCLMDLPTAVLLHRDRLGNK
MAB_2466c|M.abscessus_ATCC_199 AVDALLVRLFIRLSMALWGRRDGRAVQVIEDCIVGLPTGLLLRGRRPPDT
MSMEG_6556|M.smegmatis_MC2_155 DAQNRLVDLMIQLADDAWGRRDGAAVDTVTTCIVDLPTAILLHRNRIADP
MMAR_1142|M.marinum_M NVAELLGQVRVHHGDDTTAR--LAVANAIALRLGWRLFEPMLRSATGLDE
MUL_0905|M.ulcerans_Agy99 NVAELLGQVRVHHGDDTTAR--LAVANAIALCLGWRLFEPMLRSATGLDE
Mflv_4881|M.gilvum_PYR-GCK RLQRKLRKVLAEIAG-DHGRIDGRVDGRVAAAVMDVPAAIVRRHLRARQS
TH_0467|M.thermoresistible__bu PIITRASQRLTESFQSSWVRASEEDAGVLARAIVRLAMSYVSMPPEADHD
. * : : : .
Mvan_5831|M.vanbaalenii_PYR-1 AARDRLTAAVRAVLALTPPTSRNTPAH----------
MAV_1113|M.avium_104 AALQRLAAAVRAVLALTPPTTSTKPKPRTTLTKGAPR
MLBr_02497|M.leprae_Br4923 TARKHLHAAVRAVLEVGPPP-----------------
MAB_2466c|M.abscessus_ATCC_199 ATRSRLEAAVRAVLALNPPPANPKT------------
MSMEG_6556|M.smegmatis_MC2_155 TARRQLRAAVAAVLEVGPAPRAQRHRAQEAS------
MMAR_1142|M.marinum_M LADADLRQAVTDEVARIIEPSGRG-------------
MUL_0905|M.ulcerans_Agy99 LADADLRQAVTDEVARIIEPSGRG-------------
Mflv_4881|M.gilvum_PYR-GCK IPAAADDIVADCARALICPN-----------------
TH_0467|M.thermoresistible__bu VAHDLARLMTPFAERYGVIEEV---------------
. .