For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLDFDHLMDVAEQLAAESGPPAITIRALSEATAISNGAIYHAFGSRGGLLGHVWLRAGKRYLSFQRAAVA KALSRGVSGEAAVDAVVAAAECPATFSHEHPASARFLLRVRRDELLGAADVPAELADELRRLDDDLAHLF IELAEKVWGRRDQQAVGAIRTCVVELPTALLIDAHALHDAAALQRLAAAVRAVLALTPPTTSTKPKPRTT LTKGAPR
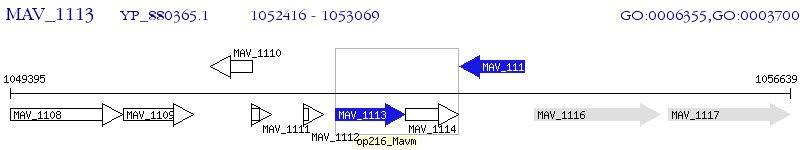
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1113 | - | - | 100% (217) | transcriptional regulator, TetR family protein |
| M. avium 104 | MAV_4715 | - | 7e-17 | 30.89% (191) | transcriptional regulator, TetR family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4881 | - | 1e-13 | 28.35% (194) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_02497 | - | 1e-32 | 49.04% (157) | hypothetical protein MLBr_02497 |
| M. abscessus ATCC 19977 | MAB_2466c | - | 8e-53 | 51.50% (200) | putative transcriptional regulator, TetR |
| M. marinum M | MMAR_1145 | - | 6e-06 | 28.24% (131) | transcriptional regulatory protein |
| M. smegmatis MC2 155 | MSMEG_6556 | - | 8e-40 | 46.35% (192) | putative transcriptional regulator |
| M. thermoresistible (build 8) | TH_2712 | - | 9e-05 | 38.64% (44) | PUTATIVE transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_0910 | - | 9e-06 | 28.24% (131) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_5831 | - | 5e-73 | 68.29% (205) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_1113|M.avium_104 ---------------------------------MLDFDHLMDVAEQLAAE
Mvan_5831|M.vanbaalenii_PYR-1 -------------MGPAITERALCYRCPVPRPRVIDLDHLMDAVEQLAAD
MLBr_02497|M.leprae_Br4923 --------------------------------------------------
MAB_2466c|M.abscessus_ATCC_199 ----------------------------MPRPRIHSVEDLLDATERIAVQ
MSMEG_6556|M.smegmatis_MC2_155 ----------------------------MPRPRMHDPQAILDVAESLAVR
MMAR_1145|M.marinum_M ----------------------MAATRTTARAGGDRRAAIIEAALNRFLS
MUL_0910|M.ulcerans_Agy99 ----------------------MAATRTTARAGGDRRAAIIEAALNRFLS
Mflv_4881|M.gilvum_PYR-GCK ----------------------------MPRPARYTVDVLLDAAAELLAA
TH_2712|M.thermoresistible__bu VSLMSADELLPARPATDVAAPTATTAPPEREDAARNRALLLDAARRLIAE
MAV_1113|M.avium_104 SGPPAITIRALSEATAISNGAIYHAFGSRGGLLGHVWLRAGKRYLSFQRA
Mvan_5831|M.vanbaalenii_PYR-1 AGPRAVTIRALSQRTAISNGAIYHAFGSRGGLLGQVWLRAARRFLSEQRS
MLBr_02497|M.leprae_Br4923 ------------------------MFASRAALLGRVWLRAGYRFLKTQTA
MAB_2466c|M.abscessus_ATCC_199 DGPAAVTVRAVSQATGISNGAIYHAFGSRGGMVGHAWLRAAQRFLDMQRD
MSMEG_6556|M.smegmatis_MC2_155 CGPAAVTIRAVGDAVGVSNGALYHEFGSRSGLLTRAWLRAVHRLQTVLTE
MMAR_1145|M.marinum_M QGIDATALRQIQRDAGVSNGSFFHHFPSKEVLAAAVYVDCVTRYQQALLK
MUL_0910|M.ulcerans_Agy99 QGIDATALRQIQRDAGVSNGSFFHHFPSKEVLTAAVYVDCVTRYQQALLK
Mflv_4881|M.gilvum_PYR-GCK EGPAAVTMSAVARSTGAPSGSVYHRFPTRAALCGELWVRTEERFQEELLE
TH_2712|M.thermoresistible__bu RGAEAVTMDDIAAAAGVGKGTLFRRFGSRAGLMLELLHDDETAFQQAFMF
* :: :
MAV_1113|M.avium_104 AVAKALSRGVSGEAAVDAVVAAAECPATFSHEHPASARFLLRVRRDELLG
Mvan_5831|M.vanbaalenii_PYR-1 AVARALADGVTPETAVEAVVAAAQCPATFLDGHPATAMYLLALPRNELLG
MLBr_02497|M.leprae_Br4923 LVDQALSASVR-RAGVEAVVAAADAPVVFAEQYPDSSRLLLTVRREELLK
MAB_2466c|M.abscessus_ATCC_199 AVDGALAESETRTAAVNAVVAAADTPAAFAERFPESSRLVLSVRREDVLG
MSMEG_6556|M.smegmatis_MC2_155 LVETA-------EPGAAAIAAAAEASIVLAERHPSSAALVLQTRRDELIG
MMAR_1145|M.marinum_M DLARY-------PDAESAVRGIVGMHLSWCTEHPEMARFLITMTEPAVHR
MUL_0910|M.ulcerans_Agy99 DLVRY-------PDAESAVRGIVGMHLSWCTEHPEMARFLITMTEPAVHR
Mflv_4881|M.gilvum_PYR-GCK ALSGD-------GDPQNRCVAGALRNMQWCRENPVEAHVLLAGPDQLDMG
TH_2712|M.thermoresistible__bu GPPPLGPG----AEPLERLLAYGRRRLQFVCDHHALLTDVSREPQ-----
. :
MAV_1113|M.avium_104 AADVPAELADELRRLDDDLAHLFIELAEKVWGR--RDQQAVGAIRTCVVE
Mvan_5831|M.vanbaalenii_PYR-1 ARDVSDELADDLRGLDGELAGLFIELAEHMWRR--RDRHAVAAIRACVVD
MLBr_02497|M.leprae_Br4923 S-FLPNEIEIELRELDALLISLLTRLAVVIWDR--KDTGAIDTITTCLMD
MAB_2466c|M.abscessus_ATCC_199 S-GIPEEVARDMAAVDALLVRLFIRLSMALWGR--RDGRAVQVIEDCIVG
MSMEG_6556|M.smegmatis_MC2_155 A-----DPPAEVKDAQNRLVDLMIQLADDAWGR--RDGAAVDTVTTCIVD
MMAR_1145|M.marinum_M A--AADELRSHNERFATAVQTWWRPHAHYGALRPLSPAHSQALWLGPAQE
MUL_0910|M.ulcerans_Agy99 A--AADELRSHNERFATAVQTWWRPHAHYGALRPLSPAHSQALWLGPAQE
Mflv_4881|M.gilvum_PYR-GCK E--WPDTVAGRRKRLQRKLRKVLAEIAG-DHGR--IDGRVDGRVAAAVMD
TH_2712|M.thermoresistible__bu -----TRFGPPMTLHHTHIRSLLAAADADGDLD---------VQADALLA
:
MAV_1113|M.avium_104 LPTALLID-AHALHDAAALQRLAAAVRAVLALTPPTTSTKPKPRTTLTKG
Mvan_5831|M.vanbaalenii_PYR-1 LPTALLLR-GQHPPDPAARDRLTAAVRAVLALTPPTSRNTPAH-------
MLBr_02497|M.leprae_Br4923 LPTAVLLH-RDRLGNKTARKHLHAAVRAVLEVGPPP--------------
MAB_2466c|M.abscessus_ATCC_199 LPTGLLLR-GRRPPDTATRSRLEAAVRAVLALNPPPANPKT---------
MSMEG_6556|M.smegmatis_MC2_155 LPTAILLH-RNRIADPTARRQLRAAVAAVLEVGPAPRAQRHRAQEAS---
MMAR_1145|M.marinum_M LVRAWLLGNLAAPPNAADAAILADAAWLCLRAGPAG--------------
MUL_0910|M.ulcerans_Agy99 LVRAWLLGNLAAPPNAADAAILADAAWLCLRAGPAG--------------
Mflv_4881|M.gilvum_PYR-GCK VPAAIVRRHLRARQSIPAAADDIVADCARALICPN---------------
TH_2712|M.thermoresistible__bu LTDPDYVRHQVTDRGVALDALADSWGDVARKLCRR---------------
: . .
MAV_1113|M.avium_104 APR
Mvan_5831|M.vanbaalenii_PYR-1 ---
MLBr_02497|M.leprae_Br4923 ---
MAB_2466c|M.abscessus_ATCC_199 ---
MSMEG_6556|M.smegmatis_MC2_155 ---
MMAR_1145|M.marinum_M ---
MUL_0910|M.ulcerans_Agy99 ---
Mflv_4881|M.gilvum_PYR-GCK ---
TH_2712|M.thermoresistible__bu ---