For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VQLLVLPRIRYADGRAPPNLGAGGVMGREAAGLNLHGEAMLDAVFEHLYNAMVITDADFDGGPFFQRCNP AFCAMTGYTEEELIGQSPRILQGPETDRKVIDELERKIRAGEFFEGSTINYRKDGLSYVVQWNISPVRDA DGAIVAYVSIQQDITARLAAERERSLLAEALNAATDPVIVMDKNFIIVFANNAFGREVGRPVDDILGHSA FELRPRTAEGLAEPQIRRVLAEGSPYHGVVSFEAPTGKRLHVDLSISSLIGYHDPNGHYVAIAPNITHMV EQQMVLQEMANMDALTGLLNRRSGQIALEIELQKAAESGAPISVAMGDIDHFKRINDRLGHAVGDDVLRS VAGSLRDGLRRNDVAIRWGGEEFLVFLPNTPLDDAVPLTERIRNHVATTPQIASDSVTMSFGVGQWRPGE SVAAFVERIDAAMYEAKSGGRDRVVSSP
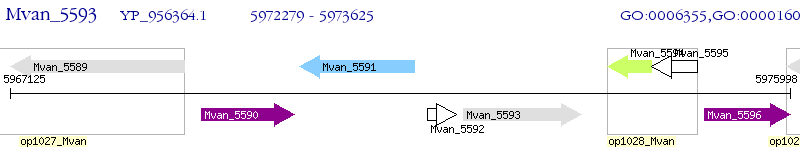
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5593 | - | - | 100% (448) | diguanylate cyclase with PAS/PAC sensor |
| M. vanbaalenii PYR-1 | Mvan_3629 | - | 2e-27 | 28.36% (402) | diguanylate cyclase with PAS/PAC sensor |
| M. vanbaalenii PYR-1 | Mvan_2109 | - | 6e-27 | 27.96% (422) | diguanylate cyclase with PAS/PAC sensor |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1389c | - | 5e-17 | 35.54% (166) | hypothetical protein Mb1389c |
| M. gilvum PYR-GCK | Mflv_2883 | - | 3e-29 | 29.68% (411) | diguanylate cyclase with PAS/PAC sensor |
| M. tuberculosis H37Rv | Rv1354c | - | 6e-17 | 35.54% (166) | hypothetical protein Rv1354c |
| M. leprae Br4923 | MLBr_00397 | - | 9e-22 | 31.67% (281) | putative transporter protein |
| M. abscessus ATCC 19977 | MAB_0124 | - | 3e-16 | 30.98% (184) | hypothetical protein MAB_0124 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3132 | - | 4e-15 | 35.15% (165) | GGDEF domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_2196 | - | 1e-16 | 34.71% (170) | sensory box/response regulator |
| M. thermoresistible (build 8) | TH_0811 | - | 1e-19 | 35.29% (170) | sensory box/response regulator |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1389c|M.bovis_AF2122/97 MCNDTATPQLEELVTTVANQLMTVDAATSAEVSQRVLAYLVEQLGVDVSF
Rv1354c|M.tuberculosis_H37Rv MCNDTATPQLEELVTTVANQLMTVDAATSAEVSQRVLAYLVEQLGVDVSF
MSMEG_2196|M.smegmatis_MC2_155 -----MSESLDVLVTSVATQLMAVEAATSVSVSQQVLAELVSFFDVDVSF
TH_0811|M.thermoresistible__bu -----VSTELDLLVTGVATKLMAADAATARDVSRRVLADLVRAFELDFSF
MAV_3132|M.avium_104 -----MSGTLDHLVTAAAAELMAATAADAAQISERVLADLVSHFGVNFGF
Mvan_5593|M.vanbaalenii_PYR-1 ------MQLLVLPRIRYADGRAPPNLGAGGVMGREAAGLNLHGEAMLDAV
Mflv_2883|M.gilvum_PYR-GCK --MRRLSTSAVVLLTFPPVFALSIVAGRATRLGGSEVALVWPAAAVAVMW
MAB_0124|M.abscessus_ATCC_1997 ----------------MGAGEMTFGQEYRFATRALTVTRNLVPLKIAIGL
MLBr_00397|M.leprae_Br4923 -----------------MDHTTKCDAEQYFQAIVTSMADGVIVVDIDGRI
:
Mb1389c|M.bovis_AF2122/97 LRHNDRDRR-------------------ATRLVAEWPPRLNIPDPDP---
Rv1354c|M.tuberculosis_H37Rv LRHNDRDRR-------------------ATRLVAEWPPRLNIPDPDP---
MSMEG_2196|M.smegmatis_MC2_155 LRHNDHRAH-------------------ATRLVAEWPPRPVDAATDP---
TH_0811|M.thermoresistible__bu LRHNDHEIR-------------------ASKLIAEWPVRENVPDPDP---
MAV_3132|M.avium_104 LRHNDHSIR-------------------ATVLVAEWPPR--NVDPDP---
Mvan_5593|M.vanbaalenii_PYR-1 FEHLYNAMV-------------------ITDADFDGGPFFQRCNPAF---
Mflv_2883|M.gilvum_PYR-GCK LLAVHHCSLRQRAIHVTLLGILAYGMNVATGATVSLSAWFVLVNVVLAVV
MAB_0124|M.abscessus_ATCC_1997 LCLSIVVLG-----------------------------------------
MLBr_00397|M.leprae_Br4923 ESINPAATR-----------------------------------------
Mb1389c|M.bovis_AF2122/97 -LRLIYFADADPVFALCEHAK-----------------------------
Rv1354c|M.tuberculosis_H37Rv -LRLIYFADADPVFALCEHAK-----------------------------
MSMEG_2196|M.smegmatis_MC2_155 -IAVVHFADADPVFAMAEHLK-----------------------------
TH_0811|M.thermoresistible__bu -IGVVHFEGADPVFALAEHAK-----------------------------
MAV_3132|M.avium_104 -LGVIYFADADSVFARVETMK-----------------------------
Mvan_5593|M.vanbaalenii_PYR-1 -CAMTGYTEEELIGQSPRILQ-----------------------------
Mflv_2883|M.gilvum_PYR-GCK TVGLLHYGRSDAVLRDPADLARLVVAVATGTMCAAVLATLYLTPVTGAPA
MAB_0124|M.abscessus_ATCC_1997 --TLIQFHPLGPHGLWPRLIHG----------------------------
MLBr_00397|M.leprae_Br4923 ---ILGLRAHDVVDMKHGHPFC----------------------------
:
Mb1389c|M.bovis_AF2122/97 -----------------------------------EPLVFRPEPATEDYQ
Rv1354c|M.tuberculosis_H37Rv -----------------------------------EPLVFRPEPATEDYQ
MSMEG_2196|M.smegmatis_MC2_155 -----------------------------------EPAVFRPEPLYDDYQ
TH_0811|M.thermoresistible__bu -----------------------------------EIHIFRPEPATDEFQ
MAV_3132|M.avium_104 -----------------------------------EPDVLRPEPANADYQ
Mvan_5593|M.vanbaalenii_PYR-1 -----------------------------------GPETDR--KVIDELE
Mflv_2883|M.gilvum_PYR-GCK WETFALFTARNGASALVGVAVWLCLRDVPWKRPRVRPAMLVEGLVVGTIV
MAB_0124|M.abscessus_ATCC_1997 --------------------------------------VLVASALVVGMC
MLBr_00397|M.leprae_Br4923 --------------------------------------FYDTDNQRVDLE
Mb1389c|M.bovis_AF2122/97 RLIEEARGVPVTSAAAVPLVSGEITTGLLGFIKFGDRKWHEAELNALMTI
Rv1354c|M.tuberculosis_H37Rv RLIEEARGVPVTSAAAVPLVSGEITTGLLGFIKFGDRKWHEAELNALMTI
MSMEG_2196|M.smegmatis_MC2_155 RTIEAGRHIPATSMACVPLLSGDVTTGVLGFVKFGDREWLPAELNALKAI
TH_0811|M.thermoresistible__bu RLIEESSGVPASTVAAVPLVSGEITTGVLGFVKYGDRQWHPDELNALKAI
MAV_3132|M.avium_104 RNIEDGTGIPEVSLAAVPLLSGDVTTGTLGFVKFGDREWLSEELNALQAI
Mvan_5593|M.vanbaalenii_PYR-1 RKIRAGEFFEGSTINYR--------KDGLSYVVQWNISPVRDADGAIVAY
Mflv_2883|M.gilvum_PYR-GCK VFVFAWSFWLNTGTPMTFVVLLPTTWVALRYSTPVSTLFVIAAANWIVFA
MAB_0124|M.abscessus_ATCC_1997 WVVLPWP-RRRVAIAFVWWADISMVVGACTLSAPASRLGALAHMGLIGVF
MLBr_00397|M.leprae_Br4923 REVMRVVRREVTTVSKVVGIDQHSGQRLWLSVNVSLLAYKAPPHSALVVS
: . :
Mb1389c|M.bovis_AF2122/97 AT------LFAQVQARVAAEARLRYLADHDDLTGLHNRRALLQHLDQRLA
Rv1354c|M.tuberculosis_H37Rv AT------LFAQVQARVAAEARLRYLADHDDLTGLHNRRALLQHLDQRLA
MSMEG_2196|M.smegmatis_MC2_155 AS------LFAQVQARIEAEERLRYLADHDHLTGLYNRRALMAHLEARLA
TH_0811|M.thermoresistible__bu AS------LFAQVQARVEAEERLRYLAEHDDLTGLHNRRALLAHLDARLA
MAV_3132|M.avium_104 AA------LFAQLQARVVAEEQIHYLAEHDELTGLLNRRALIAHLDKRLA
Mvan_5593|M.vanbaalenii_PYR-1 VS------IQQDITARLAAER---------------ERSLLAEALNAATD
Mflv_2883|M.gilvum_PYR-GCK TLSDRGPLLASDLQVRALLAQGMVFSLTVIVLALALYRDTRVRLIAELEA
MAB_0124|M.abscessus_ATCC_1997 AA------------------------------------------------
MLBr_00397|M.leprae_Br4923 FS---------DISAHHLSIERLTYEATHDCLTGLANRRFAEDQITKSLQ
Mb1389c|M.bovis_AF2122/97 PGQPG-PVAALFLDLDRLKAINDYLGHAAGDQFIHVFAQRIGDALVGESL
Rv1354c|M.tuberculosis_H37Rv PGQPG-PVAALFLDLDRLKAINDYLGHAAGDQFIHVFAQRIGDALVGESL
MSMEG_2196|M.smegmatis_MC2_155 PGQPG-PVAVMFFDLDRLKAINDYLGHTAGDAFISILAQRLQRGDDAPKL
TH_0811|M.thermoresistible__bu AGQPG-PVSALYFDLDRLKAVNDYLGHTVGDWFIRVLAERLREGAGPDDL
MAV_3132|M.avium_104 PGQPG-PVPVVFLAIDRFKVINGRLGQDAGDRFIDAFAALLREATEIPSV
Mvan_5593|M.vanbaalenii_PYR-1 P--------VIVMDKN----------------FIIVFANNAFGREVGRPV
Mflv_2883|M.gilvum_PYR-GCK SRDQA-DQDSELLGAVLDSIHDGVILVDPADRVVLRNARADDSGLVDEVV
MAB_0124|M.abscessus_ATCC_1997 --------------------------------FLLGWRVLAVHCVFATVA
MLBr_00397|M.leprae_Br4923 HDERSRLAAVLLLDLDDFKVINDSLGHDVGDAVLQTVAQRLRSAVRPDDV
.:
Mb1389c|M.bovis_AF2122/97 IARLGG------DEFVLIPASPMSADAAQPLAERLRDQLKDHVAIGGEVL
Rv1354c|M.tuberculosis_H37Rv IARLGG------DEFVLIPASPMSADAAQPLAERLRDQLKDHVAIGGEVL
MSMEG_2196|M.smegmatis_MC2_155 IARLGG------DEFVVVPDDPMSLDEATALAYRLQSVLRERVTVDGEML
TH_0811|M.thermoresistible__bu IARLGG------DEFVVVPSGPVDADAALELANRLRSRLQQQVCVDGEML
MAV_3132|M.avium_104 IARFGG------DEFGVVPAAPMDVEAAESFAHSLHERLHHPIAIDDEML
Mvan_5593|M.vanbaalenii_PYR-1 DDILGH------SAFELRPR----------TAEGLAEPQIRRVLAEGSPY
Mflv_2883|M.gilvum_PYR-GCK AAARAVGHGGADGPEPAVPHDVIVSVEDSRTLELTTTPLPRQPQFDVMAF
MAB_0124|M.abscessus_ATCC_1997 IFGLMG-----------------------WNVRLGEATWFDQYVYSAPAL
MLBr_00397|M.leprae_Br4923 VARLGG------DEFIVLLRGPLSDMNANDVAKRLHTTLSESLVVDQLTV
Mb1389c|M.bovis_AF2122/97 TRTVSIG-----------VASGTPGQHTPSDLLRRADQAALAAKHAGGDS
Rv1354c|M.tuberculosis_H37Rv TRTVSIG-----------VASGTPGQHTPSDLLRRADQAALAAKHAGGDS
MSMEG_2196|M.smegmatis_MC2_155 TRTVSIG-----------IAQGIPGKDSTSDVLNRADHAVLTAKGSGGNR
TH_0811|M.thermoresistible__bu TRTASIG-----------VAQGTPGRDTTSDLLRRADQAVLTAKSAGGNQ
MAV_3132|M.avium_104 SRTVSIG-----------VAAGIPGDESTSDLLRRVDQATRSAKAAGGNN
Mvan_5593|M.vanbaalenii_PYR-1 HGVVSFE-----------APTG-----------KRLHVDLSISSLIG---
Mflv_2883|M.gilvum_PYR-GCK RDVTEVRRNARELREARDLFAGVLQAASEQAIIGTDQDGLITVFNQGAER
MAB_0124|M.abscessus_ATCC_1997 SSVVLLP-------------------------------------------
MLBr_00397|M.leprae_Br4923 PIGASVG----------ILEVRPDDRRRAADILRDADSAMYAAKNKKQCA
. .
Mb1389c|M.bovis_AF2122/97 VAIFTADMSV------------------SGELRNDIELHLRRGIES--DA
Rv1354c|M.tuberculosis_H37Rv VAIFTADMSV------------------SGELRNDIELHLRRGIES--DA
MSMEG_2196|M.smegmatis_MC2_155 VAVFSDAMAM------------------EIDFRNDIELHLQSVIEG--GA
TH_0811|M.thermoresistible__bu ITVFTDDMSL------------------EAEFRNDIELHLQSVIQD--EA
MAV_3132|M.avium_104 VATFRPEMST------------------IDTIRNDIELYLEGMIDGGSGA
Mvan_5593|M.vanbaalenii_PYR-1 --------------------------------YHDPNGHYVAIAPN----
Mflv_2883|M.gilvum_PYR-GCK LLGWTAEEMIGRPVTDVHDLAEMIVRADELGVPPGFEAFIANVTPDDAEI
MAB_0124|M.abscessus_ATCC_1997 -------------------------------------IIIQVVVEG----
MLBr_00397|M.leprae_Br4923 VTPQQLVPFVALIALFVFFT------AAAGAKFYAPSNLLVILQQTVVLA
Mb1389c|M.bovis_AF2122/97 LRLVYLPEVDLRTGDIVGTEALVRWQHPTRGLLAPGCFIPVAESINLAGE
Rv1354c|M.tuberculosis_H37Rv LRLVYLPEVDLRTGDIVGTEALVRWQHPTRGLLAPGCFIPVAESINLAGE
MSMEG_2196|M.smegmatis_MC2_155 LVLHYLPEIDMRTGEVLAAEALVRWEHPTRGLLSPDSFIGVAESINLAGE
TH_0811|M.thermoresistible__bu LLLHFLPEVDMRTGEILATEALVRWHHPTRGLLAPDTFIGVAESVNLAGE
MAV_3132|M.avium_104 LVLHYLPEFDMRTGEILGTEALTRWQHPTLGLLMPDSFIQVAESINLGAK
Mvan_5593|M.vanbaalenii_PYR-1 --ITHMVEQQMVLQEMANMDALTGLLNRRSGQIALEIELQKAAESGAP--
Mflv_2883|M.gilvum_PYR-GCK REWTYVRRDGSHAEVSLAISQMSDADGCCVGYIGVATDITERKKAEHALA
MAB_0124|M.abscessus_ATCC_1997 -----GRRSLKAISLAAHQDPLTGLLNRRGVQVALSSLLSGQR-------
MLBr_00397|M.leprae_Br4923 IVGYGMTFVIMAGSVDLSVGSIVALTGVTAALVAAQNQFAAIVTALLVGL
: : :
Mb1389c|M.bovis_AF2122/97 LDRWVLRRACN------EFSEWQSAGLGHDALLRINVSAGQLVTGGFVDF
Rv1354c|M.tuberculosis_H37Rv LDRWVLRRACN------EFSEWQSAGLGHDALLRINVSAGQLVTGGFVDF
MSMEG_2196|M.smegmatis_MC2_155 LGRWVLRTACA------EFSRWRANGVGRNIVLRINVSPVQLVTDGFVES
TH_0811|M.thermoresistible__bu LGRWVMRTACG------EFAKWRAEGIGLDAVLRINVSPVQLVTDGFVAS
MAV_3132|M.avium_104 LGRLVMRSACA------QFGQWRSRGVGQGTVLRINVSPVQLVTAGIVDT
Mvan_5593|M.vanbaalenii_PYR-1 -----ISVAMG------DIDHFK----------RINDRLGHAVGDDVLRS
Mflv_2883|M.gilvum_PYR-GCK ESEERFRSAFDT--APMGMFMFTVTPEDAGRITRCNQALADFLGRSTSEV
MAB_0124|M.abscessus_ATCC_1997 -----------------ATRIVAVVLVDVDELKELNDRLGHDAGDESIRA
MLBr_00397|M.leprae_Br4923 AAGMVNGIVFAYGKIPSFVSTLGMLQVCRGITLMISDSSAKPMPFHGILG
. .
Mb1389c|M.bovis_AF2122/97 VADTIGQHGLDA--------------------------------------
Rv1354c|M.tuberculosis_H37Rv VADTIGQHGLDA--------------------------------------
MSMEG_2196|M.smegmatis_MC2_155 VAGIMKEFGLPR--------------------------------------
TH_0811|M.thermoresistible__bu VADIIDEFGLDG--------------------------------------
MAV_3132|M.avium_104 VAATLDEFGLDP--------------------------------------
Mvan_5593|M.vanbaalenii_PYR-1 VAGSLRDG------------------------------------------
Mflv_2883|M.gilvum_PYR-GCK PGMSVTDLGTNENADGTEGLARLLALTDGEPVSAEVRFRRWDGDTMWGAV
MAB_0124|M.abscessus_ATCC_1997 VAAMLATT------------------------------------------
MLBr_00397|M.leprae_Br4923 AMGAMPWILIVC--------------------------------------
:
Mb1389c|M.bovis_AF2122/97 -----SSVCLEITENVVVQDLHTARATLARLKEVGVHIAIDDFGTGYSAI
Rv1354c|M.tuberculosis_H37Rv -----SSVCLEITENVVVQDLHTARATLARLKEVGVHIAIDDFGTGYSAI
MSMEG_2196|M.smegmatis_MC2_155 -----GSVCLEITESVVVQDIETTRTTLTGLHNVGVQVAIDDFGTGYSVL
TH_0811|M.thermoresistible__bu -----SSVCLEITESVVVQDIKTTRITLEGLNEVGVQVAIDDFGTGYSVL
MAV_3132|M.avium_104 -----STVCLEITESVVVQDIDATRQTLFGLKDIGVQIAIDDFGTGYSVL
Mvan_5593|M.vanbaalenii_PYR-1 ----------------------------LRRNDVAIRWGGEEF-----LV
Mflv_2883|M.gilvum_PYR-GCK SASVVAAAGADPYGICLVEDITSRKRAEAELQHLALHDPLTGLANRALLV
MAB_0124|M.abscessus_ATCC_1997 ----------------------------TRASDLTARMGGDEF----MVV
MLBr_00397|M.leprae_Br4923 ------LFVTILAGILFQFTMFGRWVKAIGGNERVATLAGVPTRGIKVAI
. :
Mb1389c|M.bovis_AF2122/97 SLLQTLPIDT------------------LKIDKTFVRQLGTNTSDLVIVR
Rv1354c|M.tuberculosis_H37Rv SLLQTLPIDT------------------LKIDKTFVRQLGTNTSDLVIVR
MSMEG_2196|M.smegmatis_MC2_155 SLLKSLPVDT------------------LKIDRSFVAELGSNPGDLPIVR
TH_0811|M.thermoresistible__bu SHLKTLPVDT------------------LKIDKGFVQDLGNSPGDLAIVR
MAV_3132|M.avium_104 TYLKSLPVDS------------------LKIDKGFVQSLDTNPGDLAIVR
Mvan_5593|M.vanbaalenii_PYR-1 FLPNTPLDDA------------------VPLTERIRNHVATTP---QIAS
Mflv_2883|M.gilvum_PYR-GCK DRVEQAIADIGRHAPACVGLLYLDLDGFKPVNDTWGHAAGDEVLNAVARR
MAB_0124|M.abscessus_ATCC_1997 GFFDHADEAT-----------------------GLIERIGARRFGIGPHR
MLBr_00397|M.leprae_Br4923 FAICGLTAGLGG----------------IVLASRLGAGTPTAATGFEIDV
Mb1389c|M.bovis_AF2122/97 GIMTLAEGFQLDVVAEGVETEAAARILLDQRCYRAQGFLFSRPVPGEAMR
Rv1354c|M.tuberculosis_H37Rv GIMTLAEGFQLDVVAEGVETEAAARILLDQRCYRAQGFLFSRPVPGEAMR
MSMEG_2196|M.smegmatis_MC2_155 AVIALAGAFGLQLVAEGVETERAALTLLRHGCYRAQGFLLSKPILGSEMQ
TH_0811|M.thermoresistible__bu AIIALAEAFGLQLVAEGVETEAAALTLLRHGCHRAQGFLLSRPIPSNEMR
MAV_3132|M.avium_104 STLALADAFGLDVVAEGVETVAAAETLLSLGCHRAQGFLLSRPLDSAGME
Mvan_5593|M.vanbaalenii_PYR-1 DSVTMSFGVGQWRPGE---SVAAFVERIDAAMYEAKSGGRDRVVSSP---
Mflv_2883|M.gilvum_PYR-GCK IENSIRPGDTAARLGGDEFAVLCPVIKDSEALHGIAERIRREVRRPLRLR
MAB_0124|M.abscessus_ATCC_1997 ASVSMGSAMRSTSVAGFDFESLRRAADEELRGAKIERRNGFHAVP-----
MLBr_00397|M.leprae_Br4923 IAAVVIGGTPLTGGLGRLSGTLIGAIIISMLSNGMVFMGVGNAASQIIKG
: . .
Mb1389c|M.bovis_AF2122/97 HMLSARRLPPTCIPATDPALS-----------------------------
Rv1354c|M.tuberculosis_H37Rv HMLSARRLPPTCIPATDPALS-----------------------------
MSMEG_2196|M.smegmatis_MC2_155 TLLAKGRVPVHFSAAPRM--------------------------------
TH_0811|M.thermoresistible__bu ELLARGRIPMRFSATP----------------------------------
MAV_3132|M.avium_104 ALLIKGAIPMRFSDGGAPENQHVV--------------------------
Mvan_5593|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2883|M.gilvum_PYR-GCK AGRTYDRLSASTGAATSGGNSNAESLIAHTDRLMYRAKRNGKDRVADDLV
MAB_0124|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00397|M.leprae_Br4923 IMLAAAVFVFLQRRKIGIIK------------------------------
Mb1389c|M.bovis_AF2122/97 ----
Rv1354c|M.tuberculosis_H37Rv ----
MSMEG_2196|M.smegmatis_MC2_155 ----
TH_0811|M.thermoresistible__bu ----
MAV_3132|M.avium_104 ----
Mvan_5593|M.vanbaalenii_PYR-1 ----
Mflv_2883|M.gilvum_PYR-GCK PVEG
MAB_0124|M.abscessus_ATCC_1997 ----
MLBr_00397|M.leprae_Br4923 ----